Abstract
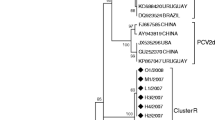
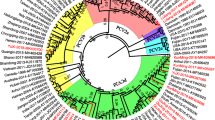
Porcine circovirus type 2 (PCV2) is the most ubiquitous viral pathogen of pigs and has persistently affected the global swine industry. Since first being identified in South Korea in 1999, the virus has undergone considerable genetic change and genotype shifts during the past two decades. These events have contributed to the coexistence of genotypes PCV2a, PCV2b, and PCV2d in Korean pig populations, which may promote viral recombination. The genotypic and phylogenetic characteristics of PCV2 strains circulating in pig herds on Jeju Island from 2019 to 2020 were the focus of this study. Genotype-specific PCR indicated that PCV2d is the dominant viral genotype and that coinfections with PCV2d and PCV2a (75%) or PCV2a and PCV2b (25%) are common in provincial pig herds. The complete genome sequences of 11 PCV2 strains, including three PCV2a, two PCV2b, and six PCV2d strains, were determined. A genomic comparison showed that all of the viruses had the highest nucleotide sequence identity to their corresponding genotypic reference strain. Notably, genetic and phylogenetic analysis revealed that one PCV2d strain, KNU-1931, exhibited nucleotide sequence variation in the ORF1 gene when compared to other PCV2d strains but showed a high degree of similarity to the PCV2b strains. Comprehensive recombination analysis suggested that KNU-1931 originated from natural recombination within ORF1 between PCV2b (the minor parent) and PCV2d (the major parent) strains. Our findings provide information about the frequency of genetic recombination between two different PCV2 genotypes circulating in the field domestically, illustrating the importance of continual intergenotypic recombination for viral fitness when multiple genotypes are present.




Similar content being viewed by others
References
Allan GM, McNeilly F, Kennedy S, Daft B, Clarke EG, Ellis JA, Haines DM, Meehan BM, Adair BM (1998) Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J Vet Diagn Invest 10:3–10
Segalés J (2012) Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology, and Laboratory diagnosis. Virus Res 164:10–19
Segalés J, Olvera A, Grau-Roma L, Charreyre C, Nauwynck H, Larsen L, Dupont K, McCullough K, Ellis J, Krakowka S, Mankertz A, Fredholm M, Fossum C, Timmusk S, Stockhofe-Zursiden N, Beattie V, Armstrong D, Grassland B, Baekbo P, Allan G (2008) PCV-2 genotype definition and nomenclature. Vet Rec 162:867–868
Ramamoorthy S, Meng XJ (2009) Porcine circoviruses: a minuscule yet paradox. Anim Health Res Rev 10:1–20
Cheung AK (2006) Rolling-circle replication of an animal circovirus genome in a theta-replicating bacterial plasmid in Escherichia coli. J Virol 80:8686–8694
Khayat R, Brunn N, Speir JA, Hardham JM, Ankenbauer RG, Schneemann A, Johnson JE (2011) The 2.3-angstrom structure of porcine circovirus 2. J Virol 85:7856–7862
Olvera A, Cortey M, Segalés J (2007) Molecular evolution of porcine circovirus type 2 genomes: phylogeny and clonality. Virology 357:175–185
He J, Cao J, Zhou N, Jin Y, Wu J, Zhou J (2013) Identification and functional analysis of the novel ORF4 protein encoded by porcine circovirus type 2. J Virol 87:1420–1429
Liu J, Chen I, Kwang J (2005) Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis. J Virol 79:8262–8274
Davies B, Wang X, Dvorak CM, Marthaler D, Murtaugh MP (2016) Diagnostic phylogenetics reveals a new Porcine circovirus 2 cluster. Virus Res 217:32–37
Harmon KM, Gauger PC, Zhang J, Piñeyro PE, Dunn DD, Chriswell AJ (2015) Whole-genome sequences of novel porcine circovirus type 2 viruses detected in swine from Mexico and the United States. Genome Announc 3:e01315–e01315
Xiao CT, Halbur PG, Opriessnig T (2015) Global molecular genetic analysis of porcine circovirus type 2 (PCV2) sequences confirms the presence of four main PCV2 genotypes and reveals a rapid increase of PCV2d. J Gen Virol 96:1830–1841
Guo LJ, Lu YH, Wei YW, Huang LP, Liu CM (2010) Porcine circovirus type 2 (PCV2): genetic variation and newly emerging genotypes in China. Virol J 7:273
Dupont K, Nielsen EO, Baekbo P, Larsen LE (2008) Genomic analysis of PCV2 isolates from danish archives and a current PMWS case-control study supports a shift in genotypes with time. Vet Microbiol 128:56–64
Franzo G, Cortey M, de Castro AM, Piovezan U, Szabo MP, Drigo M, Segalés J, Richtzenhain LJ (2015) Genetic characterisation of porcine circovirus type 2 (PCV2) strains from feral pigs in the Brazilian Pantanal: an opportunity to reconstruct the history of PCV2 evolution. Vet Microbiol 178:158–162
Lyoo YS, Kim JH, Park CK (1999) Identification of porcine circovirus with genetic variation from lymph nodes collected in pigs with PMWS. Korean J Vet Res 39:353–358
An DJ, Roh IS, Song DS, Park CK, Park BK (2007) Phylogenetic characterization of porcine circovirus type 2 in PMWS and PDNS Korean pigs between 1999 and 2006. Virus Res 129:115–122
Kim HK, Luo Y, Moon HJ, Park SJ, Keum HO, Rho S, Park BK (2009) Phylogenetic and recombination analysis of genomic sequences of PCV2 isolated in Korea. Virus Genes 39:352–358
Kwon T, Lee DU, Yoo SJ, Sang HJ, Shin JY, Lyoo YS (2017) Genotypic diversity of porcine circovirus type 2 (PCV2) and genotype shift to PCV2d in Korean pig population. Virus Res 228:24–29
Kim HR, Park YR, Lim DR, Park MJ, Park JY, Kim SH, Lee KK, Lyoo YS, Park CK (2017) Multiplex real-time polymerase chain reaction for the differential detection of porcine circovirus 2 and 3. J Virol Methods 250:11–16
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Martin DP, Murrell B, Khoosal A, Muhire B (2017) Detecting and analyzing genetic recombination using RDP4. Methods Mol Biol 1525:433–460
Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC (1999) Fulllength human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India with evidence of intersubtype recombination. J Virol 73:152–160
Kim D, Ha Y, Oh Y, Chae C (2011) Prevalence of porcine circovirus types 2a and b in pigs with and without post-weaning multi-systemic wasting syndrome. Vet J 188:115–117
Firth C, Charleston MA, Duffy S, Shapiro B, Holmes EC (2009) Insights into the evolutionary history of an emerging livestock pathogen: porcine circovirus 2. J Virol 83:12813–12821
Ramos N, Mirazo S, Castro G, Arbiza J (2013) Molecular analysis of Porcine Circovirus Type 2 strains from Uruguay: evidence for natural occurring recombination. Infect Genet Evol 19:23–31
Cai L, Han X, Hu D, Li X, Wang B, Ni J, Zhou Z, Yu X, Zhai X, Tian K (2012) A novel porcine circovirus type 2a strain, 10JS-2, with eleven-nucleotide insertions in the origin of genome replication. J Virol 86:7017
Franzo G, Cortey M, Segalés J, Hughes J, Drigo M (2016) Phylodynamic analysis of porcine circovirus type 2 reveals global waves of emerging genotypes and the circulation of recombinant forms. Mol Phylogenet Evol 100:269–280
Jiang CG, Wang G, Tu YB, Liu YG, Wang SJ, Cai XH, An TQ (2017) Genetic analysis of porcine circovirus type 2 in China. Arch Virol 162:2715–2726
Liu J, Wei C, Dai A, Lin Z, Fan K, Fan J, Liu J, Luo M, Yang X (2018) Detection of PCV2e strains in Southeast China. PeerJ 6:e4476
Neira V, Ramos N, Tapia R, Arbiza J, Neira Carrillo A, Quezada M, Ruiz Á, Bucarey S (2017) Genetic analysis of porcine circovirus type 2 from pigs affected with PMWS in Chile reveals intergenotypic recombination. Virol J 14:191
Wang H, Gu J, Xing G, Qiu X, An S, Wang Y, Zhang C, Liu C, Gong W, Tu C, Su S, Zhou J (2019) Genetic diversity of porcine circovirus type 2 in China between 1999–2017. Transbound Emerg Dis 66:599–605
Wei C, Lin Z, Dai A, Chen H, Ma Y, Li N, Wu Y, Yang X, Luo M, Liu J (2019) Emergence of a novel recombinant porcine circovirus type 2 in China: PCV2c and PCV2d recombinant. Transbound Emerg Dis 66:2496–2506
Hesse R, Kerrigan M, Rowland RR (2008) Evidence for recombination between PCV2a and PCV2b in the field. Virus Res 132:201–207
Huang Y, Shao M, Xu X, Zhang X, Du Q, Zhao X, Zhang W, Lyu Y, Tong D (2013) Evidence for different patterns of natural inter-genotype recombination between two PCV2 parental strains in the field. Virus Res 175:7886
Eddicks M, Fux R, Szikora F, Eddicks L, Majzoub-Altweck M, Hermanns W, Sutter G, Palzer A, Banholzer E, Ritzmann M (2015) Detection of a new cluster of porcine circovirus type 2b strains in domestic pigs in Germany. Vet Microbiol 176:337–343
Salgado JM, Rodríguez-Solana R, Curiel JA, de Rivas B, Munoz R, Domínguez JM (2014) Bioproduction of 4-vinylphenol from corn cob alkaline hydrolyzate in two-phase extractive fermentation using free or immobilized recombinant E. coli expressing pad gene. Enzyme Microb Technol 58–59:22–28
Seo HW, Park C, Kang I, Choi K, Jeong J, Park SJ, Chae C (2014) Genetic and antigenic characterization of a newly emerging porcine circovirus type 2b mutant first isolated in cases of vaccine failure in Korea. Arch Virol 159:3107–3111
Harding JCS, Ellis JA, McIntosh KA, Krakowka S (2010) Dual heterologous porcine circovirus genogroup 2a/2b infection induces severe disease in germ-free pigs. Vet Microbiol 145:209–219
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF- 2018R1D1A1B07040334).
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF- 2018R1D1A1B07040334).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Additional information
Handling Editor: Ana Cristina Bratanich.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jang, G., Yoo, H., Kim, Y. et al. Genetic and phylogenetic analysis of porcine circovirus type 2 on Jeju Island, South Korea, 2019–2020: evidence of a novel intergenotypic recombinant. Arch Virol 166, 1093–1102 (2021). https://doi.org/10.1007/s00705-020-04948-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04948-1