Abstract
Porcine deltacoronavirus (PDCoV) was first detected in Hong Kong and has recently spread to many countries around the world. PDCoV causes acute diarrhea and vomiting in pigs, resulting in significant economic losses in the global pork industry. In this study, a Chinese PDCoV strain, designated CHN-HG-2017, was isolated from feces of a suckling piglet with severe watery diarrhea on a farm located in central China. Subsequently, the virus was identified by an indirect immunofluorescence assay and electron microscopy. A nucleotide sequence alignment showed that the whole genome of CHN-HG-2017 is 97.6%-99.1% identical to other PDCoV strains. Analysis of potential recombination sites showed that CHN-HG-2017 is a possible recombinant originating from the strains CH/SXD1/2015 and Vietnam/HaNoi6/2015. Furthermore, the pathogenicity of this recombinant PDCoV strain was investigated in 5-day-old piglets by oral inoculation. The challenged piglets developed typical symptoms, such as vomiting, anorexia, diarrhea and lethargy, from 1 to 7 days post-inoculation (DPI). Viral shedding was detected in rectal swabs until 14 DPI in the challenged piglets. Interestingly, high titers of virus-neutralizing antibodies in sera were detected at 21 DPI. Tissues of small intestines from CHN-HG-2017-infected piglets at 4 DPI displayed significant macroscopic and microscopic lesions with clear viral antigen expression. Our analysis of the full genome sequence of a recombinant PDCoV and its virulence in suckling piglets might provide new insights into the pathogenesis of PDCoV and facilitate further investigation of this newly emerged pathogen.
Similar content being viewed by others
Introduction
Porcine deltacoronavirus (PDCoV), a member of the family Coronaviridae, order Nidovirales, is an enveloped, single-stranded positive-sense RNA virus [1]. The genome of PDCoV is 25.4 kb in length, making it the smallest genome among the known coronaviruses (CoVs) [2]. The coding region of PDCoV is flanked by short 5′ and 3′ untranslated regions (UTRs), and the gene order is 5′-UTR-ORF1a-ORF1b-S-E-M-NS6-N-NS7-3′-UTR. PDCoV encodes at least four structural proteins, including the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins, as well as two accessory proteins, NS6 and NS7 [3,4,5,6]. The S protein is responsible for receptor binding and membrane fusion and acts as the major antigen inducing neutralizing antibodies [7]. The N protein is highly conserved and plays a critical role in viral RNA encapsidation [8]. The M and E proteins are important for virion assembly and budding [9].
During a molecular surveillance study performed by Yuen and coworkers in 2012, PDCoV was first identified in swine specimens in Hong Kong [10]. Following the first detection of PDCoV in pigs with diarrhea in Ohio, USA, in February 2014, dozens of PDCoV strains were identified in the United States, Canada, South Korea, China, and Thailand [4, 5, 11,12,13,14,15,16]. Piglets infected with PDCoV are characterized by watery diarrhea, variable vomiting, and dehydration [3]. Notably, PDCoV-positive piglets are frequently co-infected with other enteric viruses, such as porcine epidemic diarrhea virus (PEDV), rotavirus, transmissible gastroenteritis coronavirus (TGEV), and porcine respiratory coronavirus (PRCV), which considerably increases the morbidity and mortality of piglets [17]. PDCoV outbreaks have been reported in many countries worldwide and have caused significant economic losses in the pork industry. Moreover, due to the unique genome replication strategy of coronavirus, PDCoV undergoes RNA mutation and recombination events at high frequency, giving rise to new variants, which makes it extremely difficult to control and eliminate [18].
In 2015, Saif and coworkers reported that a PDCoV strain named OH-FD22 caused severe atrophic enteritis accompanied by severe diarrhea and vomiting in gnotobiotic piglets [19]. Macroscopic examination revealed that the jejunum and ileum are the major targets for PDCoV infection, and the pathological lesions were similar to, but milder than, those caused by PEDV [19]. Subsequently, the pathogenicity of several American strains, including USA/IL/2014, Michigan/8977, and Ohio CVM I, was investigated in piglets [20]. These American strains exhibited pathogenic characteristics similar to those of OH-FD22. In addition, it has been reported that newborn piglets inoculated with two recent Chinese PDCoV isolates, named CHN-HN-2014 and CHN-GD-2016, developed clear clinical signs, including severe diarrhea, vomiting, and dehydration [21, 22].
In this study, we successfully isolated a high-growth PDCoV strain, hereafter referred to as CHN-HG-2017, from feces of a pig on a farm located in Huanggang, Hubei province. Analysis of the genomic sequence suggested that CHN-HG-2017 is a potential recombinant virus derived from Chinese and Vietnam PDCoV strains. Furthermore, the pathogenicity of CHN-HG-2017 in 5-day-old piglets was investigated by clinical assessment, the quantification of viral shedding and distribution, histology, and immunohistochemistry.
Materials and methods
Collection of clinical samples and treatment
Feces (n = 17) from pigs experiencing diarrhea were obtained from farms in Huanggang, Hubei province. They tested PDCoV-positive by N-gene-based RT-PCR as reported previously [12]. These clinical samples were subsequently subjected to virus isolation. Briefly, 1 g of PDCoV-positive feces was suspended in 5 ml of Dulbecco′s modified Eagle′s medium (Gibco, Grand Island, NY, USA), vortexed for 5 minutes, and centrifuged at 4000×g for 15 minutes at 4 °C. The supernatants were filtered through a 0.22-μm filter (Millipore, Billerica, MA) and stored at -80 °C for PDCoV isolation.
Virus isolation, propagation, and titration
LLC-PK1 cells (ATCC CL-101) were obtained from the American Type Culture Collection (Rockville, MD, USA) and were used for PDCoV isolation. LLC-PK1 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS, Gibco). The maintenance medium for PDCoV propagation was DMEM with 10% tryptose phosphate broth (TPB) and 10 μg of trypsin (Sigma) per ml. Cells were cultured in a 6-well plate until they reached 70-80% confluence and were then washed three times with PBS. Filtered inoculum (500 μl) in 1.5 ml of maintenance medium was added to the cell monolayers, and the cells were kept at 37 °C in 5% CO2 for 2 h. The cells were then washed three times with PBS, and 2 ml of maintenance medium was added to each well. The inoculated cells were cultured continuously at 37 °C in 5% CO2. When a cytopathic effect (CPE) was obvious in the infected cells, the plates were frozen and thawed twice. Subsequently, the supernatants were stored at -80 °C as seed stocks for plaque purification and for the next passage. The isolates were plaque-purified as described previously [20]. The virus titer was determined based on the 50% tissue culture infectious dose (TCID50) on LLC-PK1 cells in 96-well plates as described previously [20].
Immunofluorescence assay (IFA)
The N gene of CHN-HG-2017 was amplified by RT-PCR, and the PCR product was cloned into the pET28a vector. The recombinant plasmid was expressed in Escherichia coli Rosetta (DE3) under IPTG induction. Subsequently, female BALB/c mice were immunized with the purified recombinant N protein. After hybridoma cell fusion and selection, one strain of hybridoma cells that secreted an anti-N protein monoclonal antibody (mAb), named A3, was selected.
LLC-PK1 cells were seeded on sterilized coverslips placed in 24-well plates and then mock-infected or infected with plaque-purified PDCoV at a multiplicity of infection (MOI) of 0.01. At 24 h postinfection (HPI), cells were fixed with 4% paraformaldehyde for 15 min at room temperature (RT) and then permeabilized with 0.1% Triton X-100 for 10 min. Subsequently, cells were blocked with 5% skimmed milk in PBS for 1 h at room temperature and incubated with mAb A3 for 1 h. The cells were treated with fluorescein isothiocyanate (FITC)-labeled goat anti-mouse IgG secondary antibodies (Invitrogen) and then stained with 4′,6-diamidino-2-phenylindole (Invitrogen) for 15 min at room temperature. Fluorescence was examined using a fluorescence microscope (Olympus IX73, Japan).
Western blot analysis
LLC-PK1 cells were infected with PDCoV and harvested with lysis buffer (65 mM Tris-HCl [pH 6.8], 4% sodium dodecyl sulfate, 3% DL-dithiothreitol and 40% glycerol) supplemented with PMSF. Proteins isolated from the lysate were separated by 12% SDS-PAGE and then transferred to a polyvinylidene difluoride membrane. The membrane was blocked with 10% dry milk and then incubated with the PDCoV-N-specific mAb A3. After washing three times with PBST, the membrane was incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (ABclonal) for 45 min at room temperature. Signals were detected using a SuperSignal West Pico Luminal Kit (Pierce).
Electron microscopy
After inoculation with CHN-HG-2017, LLC-PK1 cells were harvested when more than 80% CPE was observed. Cells were frozen and thawed three times and then centrifuged at 10,000×g at 4 °C for 20 min. The supernatants were filtered and ultracentrifuged at 30,000×g for 2 h at 4 °C. The pellets were resuspended in PBS and centrifuged in a non-linear 30%–60% sucrose gradient at 30,000×g for 3 h at 4 °C. The virions that sedimented in the interphase between the 30% and 45% sucrose layers were diluted with sterile PBS. The sucrose was then removed by centrifugation for 1 h at 30,000×g. The virions were resuspended in PBS, stained with 2% phosphotungstic acid, and examined using a transmission electron microscope (Hitachi H-7000FA, Japan).
Phylogenetic and recombination analysis
Viral RNA was extracted from CHN-HG-2017-infected LLC-PK1 cells using TRIzol Reagent (Invitrogen, USA) according to manufacturer′s instructions. cDNA was synthesized from the extracted RNA using random primers and a Takara RNA PCR Kit (Takara, Japan). The whole genome of CHN-HG-2017 was sequenced using 26 pairs of primers reported by Lorsirigool et al. [23]. Based on the sequence that was obtained, a pair of primers (PDCoV-5′GSP, 5′- GATTACGCCAAGCTTTAGATCGCAAGGTGGGTTGGGCTCC-3′; PDCoV-3′GSP, 5′-GATTACGCCAAGCTTCGTCGTAAGACCCAACATCAAGCT-3′) was designed to verify the 5′- and 3′- terminal sequences of CHN-HG-2017 using a SMARTer RACE 5′/3′ Kit (Takara) according to manufacturer′s instructions. The sequences were combined using DNASTAR Lasergene 7.0, and the assembled genome sequence was submitted to the GenBank database under the accession number MF095123. Multiple sequence alignments and phylogenetic analysis were performed using the neighbor-joining method with MEGA 7 software (http://www.mehasoftware.net/). The aligned sequences were scanned for recombination events, using the Recombination Detection Program (RDP).
Inoculation of pigs with PDCoV strain CHN-HG-2017
Animal experiments were carried out strictly according to the Guide for the Care and Use of Animals in Research of the People′s Republic of China. All experimental procedures were approved by the Scientific Ethics Committee of Huazhong Agricultural University (HZAUSW-2018-005). Twelve 5-day-old piglets were purchased from a commercial pig farm and were randomly divided into two groups, which were housed in separated containments. Prior to PDCoV inoculation, piglets were confirmed to be negative for PDCoV, PEDV, TGEV, and rotavirus by testing rectal swabs by RT-PCR. Neutralizing antibodies against PDCoV were not detected in sera of the piglets or sows. After acclimation for 24 h, the challenged group was inoculated orally with a CHN-HG-2017 preparation with a titer of 1 × 106 TCID50/ml (10 ml per pig), while the mock-infected group was inoculated with 10 ml of maintenance medium. Clinical symptoms, including vomiting, diarrhea, lethargy, and appetite loss, were observed and recorded daily. Rectal swabs were collected daily from each piglet from the first day post-inoculation (DPI) to the end of the experiment. Swabs were submerged into 1 ml of PBS immediately after collection. Randomly selected piglets from each group (n = 3) were subjected to necropsy at 4 DPI, and prolonged PDCoV shedding was monitored in the remaining pigs until 21 DPI. At necropsy, tissues of duodenum, jejunum, ileum, cecum, colon, rectum, liver, spleen, lung, kidney, and blood were collected for analysis by real-time RT-PCR. Tissues of duodenum, jejunum, ileum, cecum, and colon were also fixed with 10% formalin for histopathology and immunohistochemistry analysis. Sera collected at 0, 4, 7, 21 DPI from piglets were tested for neutralizing antibodies against PDCoV.
Real-time RT-PCR analysis
The number of viral RNA copies in fecal swabs, sera, and tissue samples was determined by real-time RT-PCR. Briefly, fecal swab samples diluted in DMEM were homogenized and centrifuged at 6000×g for 5 min. An aliquot (200 μl) of supernatant was extracted for viral RNA isolation. One gram of various tissue samples was mechanically homogenized in 1 ml of DMEM, and 200 μl of the supernatant was used for RNA extraction. Two hundred μl of serum was also used for RNA extraction. Viral RNA was extracted using TRIzol Reagent (Invitrogen, USA) according to the manufacturer′s protocols and was treated with DNase I. One μg of total RNA was used for cDNA synthesis by reverse transcription using an iScript cDNA Synthesis Kit (Bio-Rad). cDNA amplification was performed using the iTaq Universal Probes Supermix (Bio-Rad) with primers and probes targeting the N gene of PDCoV, as described by Ma et al. [3]. The recombinant plasmid pET28a-N containing the N gene of PDCoV was serially diluted tenfold to generate a standard curve for each plate. Real-time RT-PCR was carried out using an Applied Biosystems 7500 Fast Real-Time PCR System (Life Technologies, USA). The thermal cycling parameters were as follows: 2 min at 50 °C, 10 min at 95 °C, and 40 cycles of 15 s at 95 °C and 1 min at 60 °C. The number of copies of PDCoV viral RNA in the tested samples was calculated based on the standard curve.
Neutralization assay (NA)
The presence of PDCoV-specific neutralizing antibodies in serum samples was determined using a virus neutralizing (VN) test. Briefly, LLC-PK1 cells were grown in 96-well plates until they reached 90% confluence. The CHN-HG-2017 stock was diluted in DMEM to make 200 TCID50 viral stocks. Fifty μl of the diluted virus was then mixed with an equal volume of twofold serial dilutions of individual inactivated sera in 96-well plates and incubated at 37 °C for 1 h. After washing twice with maintenance medium, LLC-PK1 cells were incubated with the mixture at 37 °C for 1 h. The mixture was then removed, and the cell monolayers were washed three times with PBS and cultured in maintenance medium at 37 °C in a 5% CO2 incubator for 2 days. The neutralization antibody titers were calculated as the reciprocal of the highest dilution of serum at which PDCoV infection was blocked.
Histology and immunohistochemistry
At necropsy, tissues of duodenum, jejunum, ileum, cecum, colon, and rectum collected from the inoculated and control groups were separated and fixed in 10% formalin for 24 h at room temperature and then dehydrated, embedded, sectioned, and mounted on glass slides. The samples were stained with hematoxylin and eosin (H&E), and the slides were examined and analyzed by conventional microscopy. Three representative villi and crypts with integrated longitudinal sections were randomly selected from each H&E-stained jejunum and ileum. Villus-height-to-crypt-depth (VH/CD) ratios were calculated using a computerized image system (NIS-Elements 3.2). Five-micron-thick sections obtained from formalin-fixed paraffin-embedded tissues were placed onto positively charged glass slides, and the slides were air dried for 30 min. The tissue sections were deparaffinized, and the slides were rinsed and then incubated with target retrieval solution (Sigma-Aldrich, USA). The sections were blocked with 1% BSA, incubated with PDCoV-N-specific monoclonal antibodies for 30 min, and then with HRP-conjugated goat anti-mouse IgG secondary antibodies (Zhongshan Biotechnology Co., Beijing, China). The samples were visualized using a substrate-chromogen reagent (3-amino-9-ethylcarbazole/H2O2, Solarbio, China). Tissues from piglets in the negative control group were used as controls.
Results
Virus isolation
Of the 17 PDCoV-positive feces inoculated onto LLC-PK1 monolayers, only one caused obvious CPE in the form of enlarged, rounded and clustered cells at 24 HPI and rapidly detached from the monolayers at 36 HPI (Fig. 1A). This PDCoV isolate, designated as CHN-HG-2017, tested positive for PDCoV and negative for other enteric viruses by RT-PCR (data not shown). CHN-HG-2017 was further passaged in LLC-PK1 cells for a total of 10 passages, indicating that the PDCoV CHN-HG-2017 strain could be stably propagated in LLC-PK1 cells.
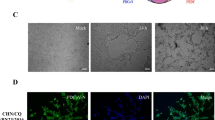
Cytopathic effect (CPE) and IFA staining of LLC-PK1 cells infected with CHN-HG-2017. (A) Infected LLC-PK1 cells at 24 HPI (a) and 36 HPI (b) and mock-infected LLC-PK1 cells at 24 HPI (d) and 36 HPI (e) are shown. PDCoV-infected (c) or mock-infected LLC-PK1 cells (f) were examined by IFA at 24 HPI using monoclonal antibodies against PDCoV N protein. (B) PDCoV-infected or mock-infected LLC-PK1 cells were analyzed at 24 HPI using a western blot assay with monoclonal antibodies against the PDCoV N protein
To confirm that virus propagation had occurred, the infectivity of the plaque-purified CHN-HG-2017 strain in LLC-PK1 cells was tested by IFA and western blot with an mAb against the PDCoV N protein. As shown in Fig. 1A, PDCoV-specific immunofluorescence was detected in most cells at 24 HPI, and the N protein was mainly localized in the cytoplasm. N protein was also detected in PDCoV-infected LLC-PK1 cells by western blot (Fig. 1B). The morphology of the PDCoV particles purified from the supernatants of CHN-HG-2017-infected LLC-PK1 cells was examined by electron microscopy (EM) using negatively stained preparations. As shown in Fig. 2, typical crown-shaped coronavirus particles with spiky surface projections and an average diameter of 80-160 nm were observed. These results clearly showed that a strain of PDCoV had been successfully isolated and identified.
Phylogenetic and recombination analysis of the genome sequence of CHN-HG-2017
The complete nucleotide sequence of the genome of CHN-HG-2017 was determined and deposited in the GenBank database under the accession number MF095123. Similar to other PDCoV strains, the complete genome sequence of CHN-HG-2017 is 25,399 nt in length, consisting of 5′ UTR-1ab-S-E-M-NS6-N-NS7-3′ UTR, and predicted transcription regulatory sequences (TRSs) were located at the 5′ end of each gene with a highly conserved core sequence of 5′ -ACCAC-3′ (Table 1). Moreover, CHN-HG-2017 shared 97.6%-99.1% nucleotide sequence identity with the other 74 PDCoV strains in the database. Phylogenetic analysis indicated that the PDCoV strains from the United States and South Korea clustered into a large clade, whereas CHN-HG-2017 clustered with other PDCoV strains detected in China since 2014. These data imply that the Chinese strains might share a more recent common ancestor with the US and South Korean strains than the ancestor of the strains from Thailand, Laos, and Vietnam (Fig. 3A). Notably, phylogenetic trees based on the S gene displayed a similar clustering pattern (Fig. 3B).
Phylogenetic analysis using nucleotide sequences of the complete genome (A) and the S gene (B) of PDCoV strains from the GenBank database. CHN-HG-2017 is indicated by a triangle. The phylogenetic tree was constructed by the neighbor-joining method in MEGA 7 (http://www.megasoftware.net). A bootstrap analysis was performed with 1000 replicates, and the bootstrap values are indicated on each branch. The scale bar indicates nucleotide substitutions per site. The branches in pink, blue, yellow and green represent a clade from the United States and South Korea; a clade from China; a clade from Thailand, Laos, and Vietnam; and the outgroup, respectively
The RNA genome of coronaviruses can undergo high-frequency recombination due to its large size [24]. We therefore analyzed the genome sequence of CHN-HG-2017 using RDP. As depicted in Fig. 4A, two PDCoV strains, Vietnam/HaNoi6/2015 (GenBank accession number: KX834351) and CH/SXD1/2015 (GenBank accession number: KT021234), might represent the parent lineages of CHN-HG-2017. Similarity plot and bootscanning analyses predicted the potential breakpoints in CHN-HG-2017 to be at nt 3153 and 4249. We therefore divided the aligned sequences into three separate segments: one from nt 1 up to the breakpoint at position 3153, another from the breakpoint at position 3153 to the breakpoint at position 4249, and the last one from the breakpoint at position 4249 to the end. A phylogenetic analysis of these segments using the maximum-likelihood algorithm suggested the CH/SXD1/2015 strain from China to be its major parent and the Vietnam/HaNoi6/2015 strain from Vietnam to be its minor parent (Fig. 4B), providing strong evidence that recombination events had occurred.
Recombination analysis of the complete genome sequence of PDCoV strain CHN-HG-2017. (A) Possible recombination events were identified using the Recombination Detection Program (RDP). Potential recombination events supported by strong P-values (< 10−10) were examined further using similarity plot and bootscan analyses implemented in Simplot 3.5.1. (B) Phylogenetic trees based on three different segments of the genome were constructed using the maximum-likelihood algorithm with the LG model with bootstrap values determined by 1000 replicates in the PhyML (version 3.0) software package
Clinical observations in pigs challenged with CHN-HG-2017
To investigate the pathogenicity of CHN-HG-2017, 5-day-old piglets were challenged with CHN-HG-2017 via oral feeding (Table 2). All piglets from the negative control groups were in good health throughout the course of study with no observed clinical signs (Fig. 5A and B). As expected, three of the six piglets in the challenged group showed diarrhea at 1 DPI, and more piglets developed varying degrees of diarrhea, vomiting, lethargy and anorexia at 2-6 DPI (Fig. 5C and D). Lethargy, anorexia and the progression to watery diarrhea were most severe at 3-6 DPI, and the piglets gradually recovered thereafter (Table 3). Notably, no PDCoV-associated deaths were observed in this study. These results demonstrated that CHN-HG-2017 is pathogenic to newborn piglets.
Fecal shedding, virus distribution, and neutralizing antibody against CHN-HG-2017
To determine the duration of fecal shedding and to monitor the changing load of CHN-HG-2017, we attempted to detect viral RNA in rectal swab samples from 1 to 21 DPI by quantitative real-time RT-PCR. The viral RNA was detected in six out of six rectal swab samples collected from the challenged group from 1 DPI to 14 DPI. The PDCoV RNA copy number in homogenized rectal swab supernatants reached a peak of over 108 copies/ml at 2-4 DPI (Fig. 6A). Notably, no PDCoV RNA was detected in samples from piglets in the negative control throughout the study.
The distribution of CHN-HG-2017 in various tissues was also examined by quantitative real-time RT-PCR in three pigs from each group sacrificed at 4 DPI. Viral RNA was detected in two of three duodenums (average 106.86 copies/g), three of three jejunums (average 108.26 copies/g), three of three ileums (average 108.4 copies/g), three of three ceca (average 108.42 copies/g), three of three colons (average 107.96 copies/g), and three of three rectums (average 106.95 copies/g). It was also detected in one of three livers (105.96 copies/g), one of three spleens (106.91 copies/g), one of three kidneys (106.9 copies/g) and one of three lungs (106.02 copies/g), but no viral RNA was detected in the blood (Fig. 6B).
To investigate when PDCoV neutralizing antibodies were detectable in the infected piglets, serum samples collected at 0, 4, 7, 21 DPI were tested for PDCoV-neutralizing antibodies. Virus-neutralizing (VN) antibodies in sera were detected as early as 4 DPI in sera of PDCoV-inoculated piglets, and the titers increased gradually and peaked at 21 DPI (Table 4). PDCoV-specific neutralizing antibodies were not observed in sera from the control piglets.
Gross pathology, histopathology, and immunohistochemistry
To determine the gross pathological and histological changes in piglets infected with CHN-HG-2017, three of the six pigs in each group were selected randomly for necropsy at 4 DPI. The small intestines, where yellow watery contents had accumulated, were clearly transparent, thin-walled, and gas-distended in the PDCoV-challenged pigs (Fig. 7A). No other lesions were observed in any other organs of the challenged groups or in any of the organs of the negative control pigs (Fig. 7B). These results showed that the small intestine is the major target of CHN-HG-2017 infection. Microscopic lesions in the small intestine were also observed by microscopy. Compared to those of the negative control piglets, the intestinal villi of the PDCoV-infected piglets were blunted and fragmented and exhibited atrophy and vacuolation, especially in the jejunum and ileum (Fig. 7C and D and Table 5). Moreover, aggregation of lymphocytes and hyperemia in the intestinal lamina propria were also observed (Fig. 7C and D), but the intestines of the negative control were normal (Fig. 7E and F). Consistent with the histopathological results, PDCoV antigen was detected by immunohistochemical staining in the cytoplasm of the infected villous enterocytes (Fig. 7G and H). No PDCoV antigen was detected in the negative controls (Fig. 7I and J). These results show that CHN-HG-2017 infection can cause intestinal lesions in newborn piglets, especially in the jejunum and ileum.
Intestinal lesions in a PDCoV-challenged pig. (A) Macroscopic lesions of a PDCoV-challenged piglet at 4 DPI. (B) Macroscopic picture of a negative control piglet at 4 DPI. (C, D) H&E-stained jejunum (C) and ileum (D) tissue section of a PDCoV-challenged piglet. (E, F) H&E-stained jejunum (E) and ileum (F) tissue section of a negative control pig. (G, H) Immunohistochemically stained jejunum (G) and ileum(H) tissue section of a PDCoV challenged piglet. (I, J) Immunohistochemically stained jejunum (I) and ileum (J) tissue section of a negative control pig. Scale bars are shown in each picture
Discussion
PDCoV is a newly emerged enteropathogenic coronavirus that causes diarrhea, dehydration and death in neonatal piglets. Since PDCoV was first reported in Hong Kong, it has spread to many countries around the world, leading to significant economic losses in the pork industry. In mainland China, the prevalence of PDCoV in pigs was reported to be 33.71% in Jiangxi province [17], 23.4% in Guangdong province [15], and 10.2% in Hebei province [25]. Although the prevalence of PDCoV in China has been widely reported, only a few PDCoV strains could be successfully isolated in cultured cells until now.
In the present study, feces from PDCoV-infected pigs on farms located in central China were used to isolate PDCoV in LLC-PK1 cells. Typical CPE was observed in infected LLC-PK1 cells at 24 h after inoculation. Following plaque purification, the PDCoV strain was confirmed by IFA and electron microscopy. Notably, only one PDCoV strain was isolated from a total of 17 PDCoV-positive feces samples, indicating that the success rate for virus isolation was still relatively low. This low rate of PDCoV isolation might be attributed to a small amount of infectious virus in the fecal samples collected for virus isolation. In addition, the time at which the sample is collected might also contribute to the success of PDCoV isolation.
To characterize the virus isolate, the complete genome of CHN-HG-2017 was sequenced and analyzed. While the genome organization is similar to those of other reported PDCoVs, the genome size of 25,399 nt is somewhat different from the others. Moreover, we identified predicted TRSs at the 5′ end of each gene of CHN-HG-2017, each with a highly conserved core sequence of 5′-ACCAC-3′, which appears to be unique to members of the genus Deltacoronavirus. Multiple sequence alignment and phylogenetic analysis showed that all of the known PDCoV strains share a high level of nucleotide sequence identities. However, CHN-HG-2017 was most closely related to other Chinese PDCoV strains. Our phylogenetic analysis suggests that PDCoV strains from China, the United States and South Korea are clustered in the same clade, whereas the PDCoV strains from Thailand, Laos, and Vietnam are clustered in another clade.
Like other coronaviruses, PDCoV has the potential to undergo recombination at a very high frequency. In this study, we employed the Recombination Detection Program (RDP) to analyze the genome of CHN-HG-2017 in search of any recombined fragments. Intriguingly, CHN-HG-2017 was found to be a possible recombinant virus derived from parental strains from China and Vietnam. The major parent and minor parent were identified as strain SXD1 from China and strain HaNoi6 from Vietnam, respectively. The potential breakpoints in CHN-HG-2017 were located at nt 3153 and 4249 in the nsp3 region. Deletion mutations in the nsp3 region in PDCoV have been reported recently [6, 26], suggesting that nsp3 which encodes a papain-like protease, may be a hotspot for genetic modification. To our knowledge, this is the first time that a recombination event has been identified in the Nsp3 region of PDCoV. The emergence of recombinant strains from China and Vietnam will probably bring new challenges to the prevention and control of PDCoV.
To investigate the pathogenicity of this putative recombinant strain, 5-day-old piglets were inoculated orally with CHN-HG-2017. As expected, severe diarrhea with vomiting and lethargy in piglets was observed from 1 to 6 DPI, suggesting that CHN-HG-2017 is pathogenic to newborn piglets. The viral RNA distribution in various tissues was also examined. Viral RNA levels were higher in intestines than in other tissues. Previously, it was reported that coronavirus HKU15 may be able to spread through the respiratory route, in addition to fecal-oral transmission [27]. In line with other reports, viral RNA of CHN-HG-2017 was also detected in the lungs of some animals, albeit with no gross lesions observed, suggesting that PDCoV may also cause respiratory infection.
It is noteworthy that we only observed microscopic lesions in the jejunum and ileum in piglets challenged with CHN-HG-2017. In striking contrast, microscopic lesions were observed in all sections of small intestine infected with PEDV [28]. Moreover, unlike PEDV, PDCoV infection usually does not result in the death of newborn piglets despite the fact that these animals exhibit lethargy and anorexia and have watery diarrhea. These results indicated that the virulence of PDCoV is milder than that of PEDV.
Recently, it was shown that VN antibodies could be detected in 14-day-old piglets inoculated with PDCoV OH-FD22 at about 7 DPI and that VN antibody titers increased gradually and remained high at the end of the experiment [29]. However, to our knowledge, there are currently no reports on the development of PDCoV-specific antibodies in sera of pigs infected with Chinese PDCoV. Our study showed that serum PDCoV VN antibodies could be detected as early as 4 DPI in PDCoV-infected piglets. VN antibody titers increased and remained high until 21 DPI, when all pigs in the challenged groups had completely recuperated.
In conclusion, we successfully isolated, characterized and obtained a high titer of CHN-HG-2017 in cultured cells. We showed that CHN-HG-2017 is probably a recombinant virus derived from CH/SXD1/2015 and Vietnam/HaNoi6/2015. Pathogenicity testing of CHN-HG-2017 revealed that this virus is still pathogenic in piglets despite multiple passages in cultured cells. High levels of serum VN antibodies were also detected in sera of infected animals. Not only will our findings be useful for understanding the molecular epidemiology and pathogenesis of PDCoV, but they also provide insights for the development of effective vaccines against this important pathogen.
References
Lee S, Lee C (2014) Complete genome characterization of Korean porcine deltacoronavirus strain KOR/KNU14-04/2014. Genome Announcements 2:e01191-01114
Woo PCY, Lau SKP, Lam CSF, Lai KKY, Huang Y et al (2009) Comparative analysis of complete genome sequences of three Avian coronaviruses reveals a novel group 3c coronavirus. J Virol 83:908–917
Ma Y, Zhang Y, Liang X, Lou F, Oglesbee M et al (2015) Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio 6: e00064-00015
Marthaler D, Jiang Y, Collins J, Rossow K (2014) Complete genome sequence of strain SDCV/USA/Illinois121/2014, a porcine deltacoronavirus from the United States. Genome Announc 2:e00218-00214
Li G, Chen Q, Harmon KM, Yoon K-J, Schwartz KJ et al (2014) Full-length genome sequence of porcine deltacoronavirus strain USA/IA/2014/8734. Genome Announc 2:e00278-00214
Wang Y-W, Yue H, Fang W, Huang Y-W (2015) Complete genome sequence of porcine deltacoronavirus strain CH/Sichuan/S27/2012 from mainland China. Genome Announc 3:e00945-00915
Godet M, Grosclaude J, Delmas B, Laude H (1994) Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J Virol 68:8008–8016
Zúñiga S, Sola I, Moreno JL, Sabella P, Plana-Durán J et al (2007) Coronavirus nucleocapsid protein is an RNA chaperone. Virology 357:215–227
Vennema H, Godeke GJ, Rossen JW, Voorhout WF, Horzinek MC et al (1996) Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J 15:2020–2028
Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, et al. (2012) Discovery of seven novel mammalian and avian coronaviruses in Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronaviruses as the gene source of Gammacoronavirus and Deltacoronavirus. JVI 06540–06511
Marthaler D, Raymond L, Jiang Y, Collins J, Rossow K et al (2014) Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg Infect Dis 20:1347
Wang L, Byrum B, Zhang Y (2014) Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg Infect Dis 20:1227
Wang L, Byrum B, Zhang Y (2014) Porcine coronavirus HKU15 detected in 9 US states, 2014. Emerg Infect Dis 20:1594–1595
Wang L, Zhang Y, Byrum B (2014) Complete genome sequence of porcine coronavirus HKU15 strain IN2847 from the United States. Genome Announc 2:e00291-00214
Chen F, Zhu Y, Wu M, Ku X, Yao L et al (2015) Full-length genome characterization of Chinese porcine deltacoronavirus strain CH/SXD1/2015. Genome Announc 3:e01284-01215
Janetanakit T, Lumyai M, Bunpapong N, Boonyapisitsopa S, Chaiyawong S et al (2016) Porcine deltacoronavirus, Thailand, 2015. Emerg Infect Dis 22:757
Song D, Zhou X, Peng Q, Chen Y, Zhang F et al (2015) Newly emerged porcine deltacoronavirus associated with diarrhoea in Swine in China: identification, prevalence and full-length genome sequence analysis. Transbound Emerg Dis 62:575–580
Makino S, Keck JG, Stohlman SA, Lai M (1986) High-frequency RNA recombination of murine coronaviruses. J Virol 57:729–737
Jung K, Hu H, Eyerly B, Lu Z, Chepngeno J et al (2015) Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg Infect Dis 21:650
Hu H, Jung K, Vlasova AN, Chepngeno J, Lu Z et al (2015) Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J Clin Microbiol 53:1537–1548
Dong N, Fang L, Yang H, Liu H, Du T et al (2016) Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet Microbiol 196:98–106
Xu Z, Zhong H, Zhou Q, Du Y, Chen L et al (2018) A highly pathogenic strain of porcine deltacoronavirus caused watery diarrhea in newborn piglets. Virol Sin 33:131–141
Lorsirigool A, Saeng-chuto K, Madapong A, Temeeyasen G, Tripipat T et al (2017) The genetic diversity and complete genome analysis of two novel porcine deltacoronavirus isolates in Thailand in 2015. Virus Genes 53:240–248
Masters PS (2006) The molecular biology of coronaviruses. Adv Virus Res. Academic Press, pp 193–292
Liu BJ, Zuo YZ, Gu WY, Luo SX, Shi QK, et al. (2018) Isolation and phylogenetic analysis of porcine deltacoronavirus from pigs with diarrhoea in Hebei province, China. Transbound Emerg Dis
Liu C, Zhang X, Zhang Z, Chen R, Zhang Z et al (2017) Complete genome characterization of novel Chinese Porcine deltacoronavirus strain SD. Genome Announc 5:e00930-00917
Woo PCY, Lau SKP, Tsang C-C, Lau CCY, Wong P-C et al (2017) Coronavirus HKU15 in respiratory tract of pigs and first discovery of coronavirus quasispecies in 5′-untranslated region. Emerg Microbes Infect 6:e53
Madson DM, Magstadt DR, Arruda PHE, Hoang H, Sun D et al (2014) Pathogenesis of porcine epidemic diarrhea virus isolate (US/Iowa/18984/2013) in 3-week-old weaned pigs. Vet Microbiol 174:60–68
Hu H, Jung K, Vlasova AN, Saif LJ (2016) Experimental infection of gnotobiotic pigs with the cell-culture-adapted porcine deltacoronavirus strain OH-FD22. Adv Virol 161:1–14
Acknowledgements
This study was supported by the National Key Research and Development Program of China (2018YFD0501102), the China Agriculture Research System (CARS-35), and the Applied Basic Research Project of Wuhan (Grant no. 2017020201010227).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have declared no conflict of interest.
Ethical approval
Necessary approvals from the Scientific Ethics Committee of Huazhong Agricultural University (HZAUSW-2018-005) were obtained prior to the start of animal experiments. This article does not contain any studies with human participants performed by any of the authors.
Additional information
Handling Editor: William G Dundon.
Rights and permissions
About this article
Cite this article
Zhang, MJ., Liu, DJ., Liu, XL. et al. Genomic characterization and pathogenicity of porcine deltacoronavirus strain CHN-HG-2017 from China. Arch Virol 164, 413–425 (2019). https://doi.org/10.1007/s00705-018-4081-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-4081-6