Abstract
Infection with shrimp hemocyte iridescent virus (SHIV), a new virus of the family Iridoviridae isolated in China, results in a high mortality rate in white leg shrimp (Litopenaeus vannamei). The complete genome sequence of SHIV was determined and analyzed in this study. The genomic DNA was 165,809 bp long with 34.6% G+C content and 170 open reading frames (ORFs). Dotplot analysis showed that the longest repetitive region was 320 bp in length, including 11 repetitions of an 18-bp sequence and 3.1 repetitions of a 39-bp sequence. Two phylogenetic trees were constructed based on 27 or 16 concatenated sequences of proteins encoded by genes that are conserved between SHIV homologous and other iridescent viruses. The results of this study, suggest that SHIV should be considered a member of the proposed new genus “Xiairidovirus”.
Similar content being viewed by others
The Iridoviridae are a family of large, icosahedral viruses with double-stranded DNA genomes ranging in size from 103 to 220 kbp. The iridescent viruses of the subfamily Alphairidovirinae infect ectothermic vertebrates (bony fish, amphibians, and reptiles), whereas members of the subfamily Betairidovirinae mainly infect insects and crustaceans [1]. The iridescent viruses contain circularly permutated and terminally redundant double-stranded genomes [2, 3]. To date, the complete genomes of 39 iridescent viruses have been sequenced (Supplemental File 1). Twenty of these are members of the genus Ranavirus, seven are members of the genus Megalocytivirus, three are members of the genus Lymphocystivirus, seven are members of the genus Iridovirus, and one is a member of the genus Chloriridovirus.
Shrimp hemocyte iridescent virus (SHIV) caused severe disease and high mortality in farmed Litopenaeus vannamei in December of 2014 in Zhejiang Province in China and was isolated and identified by Qiu et al. in 2017 [4]. PCR test results showed that farmed L. vannamei, Fenneropenaeus chinensis, and Macrobrachium rosenbergii were SHIV positive, and the detection rate was 15.8% in farmed shrimp samples collected from 2014 to 2016, indicating that SHIV is a new threat to the shrimp culture industry in China. SHIV exhibited a typical icosahedral structure with a mean diameter of 158.6 ± 12.5 nm (n = 30). Phylogenetic analysis using amino acid sequences of the major capsid protein (MCP) and ATPase indicated that it belongs to the proposed genus “Xiairidovirus”. [4]. Since only the MCP and ATPase sequences of SHIV were determined previously (accession numbers KY681039 and KY681040), the relationship of the complete genome sequence SHIV to those of other iridescent viruses has not yet been examined.
The nucleotide sequence of the SHIV genome was determined by the viral metagenomics sequencing method described by Qiu et al. [4], using an Illumina HiSeq 2500 (PE125) instrument. Gaps between assembled fragments were filled and the complete genome sequence was verified using the primer walking method (Fig. 1a). Information about the primers used in this study is shown in Supplemental File 2. DNA sequencing reads corresponding to SHIV sequences were mapped across the length of the genome (Fig. 1b). The sequencing coverage was 100%, and the fold coverage per base ranged from 101 to 118,328, with an average of 6364.7, indicating that the sequencing data completely covered the SHIV genome. The sequencing depth was uneven in distribution, possibly because of preferences in sequencing and amplification. The complete genome sequence of SHIV obtained from the original tissue has been deposited in the GenBank database (accession number MF599468).
a. Circular map of the 165,809-bp SHIV genome. The outer scale is numbered clockwise in bp. Circles 1 and 2 (from outside to inside) show ORFs on the forward and reverse strand, respectively). Circle 3 represents the G+C content along the genome sequence. Peaks toward the outside indicate that the G+C content is greater than 50%; peaks toward the inside indicate that the G+C content is lower than 50%; the height is proportional to the content). b. Sequence coverage of SHIV genomic sequences. Illumina reads were mapped to the assembled SHIV sequence using the software BWA 0.7.8. Sequence coverage was calculated according to the number of times each base mapped to the SHIV genome
The SHIV genome consists of a double-stranded DNA molecule of 165,809 bp. A comparison with other members of the family Iridoviridae showed that the SHIV genome is larger than those of members of the genera Ranavirus (103,681-140,131 bp) and Megalocytivirus (110,104-112,636 bp) but smaller than those of members of the genera Iridovirus (197,693-220,222 bp) and the chloriridovirus IIV-3 (191,100 bp). Comparison with the three sequenced members of the genus Lymphocystivirus showed that the SHIV genome is much larger than that of LCDV-1 (102,653 bp) but smaller than those of LCDV-Sa (208,501 bp) and LCDV-C (186,250 bp) (Supplemental File 1). The G+C ratio of the SHIV genome is 34.6%, which is higher than that of the members of the genera Iridovirus (28.1-30.9%), except for IIV-31 (35.09%), and Lymphocystis (27.3-33.0%) but lower than that of the members of the genera Megalocytivirus (53.0-55.0%) and Ranavirus (48.6-57.0%) and the chloriridovirus IIV-3 (47.89%) (Supplemental File 1).
To date, five iridescent viruses were found in crustaceans [4]. However, IIV-31, which was isolated from the pill bug, Armadillidium vulgare [5, 6], is the only one for which a complete genome sequence is available. IIV-31 has the largest genome size (220,222 bp) among all sequenced viruses of the family Iridoviridae and is much larger than SHIV, but the G+C content of IIV-31 (35.1%) is relatively close to that of SHIV.
We did a dotplot analysis of the SHIV genome using Genious 10.2.2. Like other iridescent viruses, the SHIV genome was found to contain numerous repetitive sequences. The longest repetitive region was identified at nt position 27,052 to 27,371 (ORF 29R). The 320-bp repetitive sequence consists of 11 repetitions of CAACCACCACAGAACCTC and 3.1 repetitions of CAACCGAATCCATTGAAATGGAAAATGTAACATCCACCA. There were some direct, inverted and palindromic repetitive sequences located at positions 113,519 to 113,693 (non-coding region) and 114,420 to 114,564 (non-coding region). The biological function of these repetitive sequences remains to be determined.
One hundred seventy open reading frames (ORFs) encoding proteins ranging from 21 to 1,167 amino acids were identified in the SHIV genome using the software GeneMark (http://topaz.gatech.edu/GeneMark/). Of the 170 ORFs, 102 were in the forward orientation (R), and 68 were in the reverse orientation (L) (Fig. 1a and Supplement File 3). The number of ORFs predicted for SHIV (170) is fewer than that of most members of the genus Iridovirus (167-468) and all three sequenced members of the genus Lymphocystivirus (183-240) but more than that of members of the genera Ranavirus (91-162), Megalocytivirus (108-135), and Chloriridovirus (126) (Supplemental File 1). There are 11 pairs of overlapping ORFs in the SHIV genome: 3L/4L, 6L/7R, 24R/25R, 34L/35R, 41R/42R, 56R/57R, 68R/69R, 84L/85L, 93R/94L, 148L/149L, and 169R/170R. Nine of these 11 pairs of ORFs have an overlap of 1-7 bp, which is similar to what has been reported for TRBIV, ISKNV, OSGIV, ATV, and SGIV [7].
Analysis using Blast2GO to identify putative proteins revealed that 63 ORFs (37.1%) had been annotated with known functions, including enzymes and structural proteins including the ATPase, DNA polymerase and major capsid protein. A BLASTp analysis using sequences from members of the family Iridoviridae showed that 22 ORFs were most similar to genes of IIV-31, the only iridescent virus found in crustaceans for which a genome sequence is available. Nineteen ORFs were most similar to the genes of IIV-6 and LCDV-Sa, which were isolated from the stem-boring lepidopterans Chilo suppressalis [8] and Sparus aurata [9], respectively. The results show that SHIV might be comparatively closely related to IIV-31, IIV-6, and LCDV-Sa.
Of the 170 ORFs in the SHIV genome, 27 (Supplemental File 3) showed sequence similarity to regions of 15 viral genomes representing all five genera of the family Iridoviridae, suggesting that these 27 ORFs are conserved and can be used for phylogenetic analysis of SHIV with other iridescent viruses (Supplemental File 4). In a previous report, Eaton et al. [10] reported 26 conserved genes that are present in every iridescent virus sequenced up to the time of the report. Like the 26 genes reported by Eaton et al. [10], the majority of the 27 conserved genes identified in this study (24 of 27 ORFs) have a predicted function based on sequence similarity of their encoded proteins to other characterized proteins. Some of these genes, including those encoding the membrane protein, RNA polymerase, NTPase, RAD2, ICP-46, MCP, deoxynucleoside kinase, ribonuclease III, DNA polymerase, phosphotransferase, and tyrosine kinase, are present among the 26 genes reported previously as well as the 27 genes identified in this study.
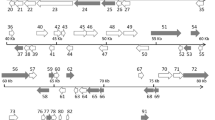
Twenty-seven conserved SHIV ORFs and the orthologous genes from 15 completely sequenced iridescent viruses of five genera were used to generate a phylogenetic tree (Fig. 2a). We further selected 16 of the 27 conserved genes and the orthologous genes from 34 completely sequenced iridescent viruses to establish a more complete phylogenetic tree of the family Iridoviridae (Fig. 2b). Both phylogenetic trees showed that the members of the five genera of the family Iridoviridae formed separate branches, and there was high bootstrap support (100%) for SHIV forming a new branch in the subfamily Betairidovirinae. Qiu et al. [4] suggested assigning SHIV to a new genus, tentatively named “Xiairidovirus” that includes iridescent viruses that infect shrimp, lobster or crayfish. The findings in this study support this proposal.
Concatenated phylogenetic tree of iridescent viruses. a. Twenty-seven conserved genes from 15 other completely sequenced iridescent viruses were rearranged as continuous amino acid sequences with the same order as in SHIV. A multiple sequence alignment was carried out using MUSCLE. The tree was reconstructed by the maximum-likelihood method using MEGA 5.0 and the numbers indicate percentage bootstrap support. b. Sixteen of the 27 conserved genes from 34 completely sequenced iridescent viruses underwent the same process to establish a tree that includes most of the completely sequenced viruses of the family Iridoviridae. Percentage bootstrap values (1000 replicates) are shown. Bar, expected nucleotide substitutions per site
In the two phylogenetic trees, four members of the genus Iridovirus (IIV-9, IIV-22, IIV-25, and IIV-30) and one member of the genus Chloriridovirus (IIV-3) were in the same branch. Similarly, phylogenetic analysis of proteins encoded by IIV3, IIV6, and IIV9 revealed that IIV9 is more closely related to IIV3 than to IIV6 [11,12,13]. This was also true of IIV22, IIV22A, IIV25, and IIV30, four close relatives of IIV9 [14,15,16,17], indicating that some members of the genus Iridovirus are more closely related to members of the genus Chloriridovirus than to other iridoviruses from insects [18]. Only two species in the genus Iridovirus, Invertebrate iridescent virus 1 and Invertebrate iridescent virus 6, have been recognized by the International Committee on Taxonomy of Viruses (ICTV). More biological and genomic data are needed before it can be determined whether other related viruses belong to the genus Iridovirus [18]. Based on the phylogenetic analysis results available to date, we suggested that IIV-9, IIV-22, IIV-25, and IIV-30 be assigned to the genus Chloriridovirus. Our study provides new information for future studies of the epidemiology of SHIV and the taxonomy and evolution of the family Iridoviridae.
References
Gregory CV, Hick P, Ince IA, Jancovich JK, Marschang R, Qin Q, Subramaniam K, Waltzek TB, Whittington R, Williams T, Zhang QY, Ictv Report Consortium (2017) Ictv virus taxonomy profile: iridoviridae. J Gen Virol. https://doi.org/10.1099/jgv.0.000818
Chinchar VG, Hyatt A, Miyazaki T, Williams T (2009) Family iridoviridae: poor viral relations no longer. Curr Top Microbiol Immunol 328:123–170
Chinchar VG, Yu KH, Jancovich JK (2011) The molecular biology of frog virus 3 and other iridoviruses infecting cold-blooded vertebrates. Viruses 3:1959–1985. https://doi.org/10.3390/v3101959
Qiu L, Chen MM, Wan XY, Li C, Zhang QL, Wang RY, Cheng DY, Dong X, Yang B, Wang XH, Xiang JH, Huang J (2017) Characterization of a new member of Iridoviridae, Shrimp hemocyte iridescent virus (SHIV), found in white leg shrimp (Litopenaeus vannamei). Sci Rep 7(1):11834. https://doi.org/10.1038/s41598-017-10738-8
Federici B (1980) Isolation of an iridovirus from two terrestrial isopods, the pill bug, Armadillidium vulgare, and the sow bug, Porcellio dilatatus. J Invertebr Pathol 36:373–381
Williams T (1994) Comparative studies of iridoviruses: further support for a new classification. Virus Res 33:99–121
Shi CY, Jia KT, Yang B, Huang J (2010) Complete genome sequence of a Megalocytivirus (family Iridoviridae) associated with turbot mortality in China. Virol J 7:1–9. https://doi.org/10.1186/1743-422X-7-159
Fukaya M, Nasu S (2008) A Chilo iridescent virus (CIV) from the rice stem borer, Chilo suppressalis Walker (Lepidoptera: Pyralidae). Appl Entomol Zool 1:69–72
López-Bueno A, Mavian C, Labella AM, Castro D, Borrego JJ, Alcami A, Alejo A (2016) Concurrence of iridovirus, polyomavirus and a unique member of a new group of fish papillomaviruses in lymphocystis disease affected gilthead seabream. J Virol 90:8768–8779. https://doi.org/10.1128/JVI.01369-16
Eaton HE, Metcalf J, Penny E, Tcherepanov V, Upton C, Brunetti CR (2007) Comparative genomic analysis of the family Iridoviridae: re-annotating and defining the core set of iridovirus genes. Virol J 4:11. https://doi.org/10.1186/1743-422X-4-11
Wong CK, Young VL, Kleffmann T, Ward VK (2011) Genomic and proteomic analysis of invertebrate iridovirus type 9. J Virol 85:7900–7911. https://doi.org/10.1128/JVI.00645-11
Lei XY, Ou T, Zhu RL, Zhang QY (2012) Sequencing and analysis of the complete genome of Rana grylio virus (RGV). Arch Virol 157:1559–1564. https://doi.org/10.1007/s00705-012-1316-9
Huang Y, Li S, Zhao Q, Pei G, An X, Guo X, Zhou H, Zhang Z, Zhang J, Tong Y (2015) Isolation and characterization of a novel invertebrate iridovirus from adult Anopheles minimus (AMIV) in china. J Invertebr Pathol 127:1–5. https://doi.org/10.1016/j.jip.2015.01.007
Piégu B, Guizard S, Spears T, Cruaud C, Couloux A, Bideshi DK, Federici BA, Bigot Y (2013) Complete genome sequence of invertebrate iridescent virus 22 isolated from a blackfly larva. J Gen Virol 94:2112–2116. https://doi.org/10.1099/vir.0.054213-0
Piégu B, Guizard S, Spears T, Cruaud C, Couloux A, Bideshi DK, Federici BA, Bigot Y (2013) Complete genome sequence of invertebrate iridovirus IIV22A, a variant of IIV22, isolated originally from a blackfly larvae. Stand Genom Sci 9:940–947. https://doi.org/10.4056/sigs.5059132
Piégu B, Guizard S, Spears T, Cruaud C, Couloux A, Bideshi DK, Federici BA, Bigot Y (2013) Complete genome sequence of invertebrate iridovirus IIV-25 isolated from a blackfly larva. Arch Virol 159:1181–1185. https://doi.org/10.1007/s00705-013-1918-x
Piégu B, Guizard S, Spears T, Cruaud C, Couloux A, Bideshi DK, Federici BA, Bigot Y (2013) Complete genome sequence of invertebrate iridovirus IIV30 isolated from the corn earworm, Helicoverpa zea. J Invertebr Pathol 116:43–47. https://doi.org/10.1016/j.jip.2013.12.007
Piégu B, Guizard S, Spears T, Cruaud C, Couloux A, Bideshi DK, Federici BA, Bigot Y (2014) Genome sequence of a crustacean iridovirus, iiv31, isolated from the pill bug, Armadillidium vulgare. J Gen Virol 95:1585–1590. https://doi.org/10.1099/vir.0.066076-0
Acknowledgements
This work was supported by projects under China Agriculture Research System (CARS-47), China ASEAN Maritime Cooperation Fund Project (2016-2018), the Program of Introducing Internationally Advanced Agricultural Science and Technology (948: 2016-X56), the Project of the Aoshan Science and Technology Innovation Program of Qingdao National Laboratory for Marine Science and Technology (2015ASKJ02), and the Construction Programme for Distinguished Taishan Scholars of Shandong Province of China. The authors thank Ms. Fang Ma and Luo-Luo Zhu of Shanghai Ocean University. We also thank Novogene Co., LTD for the contribution to metagenomics sequencing in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that we have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Chan-Shing Lin.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Qiu, L., Chen, MM., Wang, RY. et al. Complete genome sequence of shrimp hemocyte iridescent virus (SHIV) isolated from white leg shrimp, Litopenaeus vannamei . Arch Virol 163, 781–785 (2018). https://doi.org/10.1007/s00705-017-3642-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3642-4