Abstract
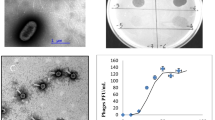
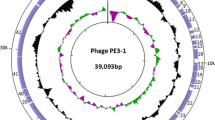
A novel low-temperature Escherichia coli phage vB_EcoS_NBD2 was isolated in Lithuania from agricultural soil. With an optimum temperature for plating around 20 °C, vB_EcoS_NBD2 efficiently produced plaques on Escherichia coli NovaBlue (DE3) at a temperature range of 10–30 °C, yet failed to plate at temperatures above 35 °C. Phage vB_EcoS_NBD2 virions have a siphoviral morphology with an isometric head (65 nm in diameter), and a non-contractile flexible tail (170 nm). The 51,802-bp genome of vB_EcoS_NBD2 has a G + C content of 49.8%, and contains 87 probable protein-encoding genes as well as 1 gene for tRNASer. Comparative sequence analysis revealed that 22 vB_EcoS_NBD2 ORFs encode unique proteins that have no reliable identity to database entries. Based on homology to biologically defined proteins and/or proteomics analysis, 36 vB_EcoS_NBD2 ORFs were given a putative functional annotation, including 20 genes coding for morphogenesis-related proteins and 13 associated with DNA metabolism. Phylogenetic analysis revealed that vB_EcoS_NBD2 belongs to the subfamily Tunavirinae, but cannot be assigned to any genus currently recognized by ICTV.



Similar content being viewed by others
References
Salmond GP, Fineran PC (2015) A century of the phage: past, present and future. Nat Rev Microbiol 13:777–786
Clokie MR, Millard AD, Letarov AV, Heaphy S (2011) Phages in nature. Bacteriophage 1:31–45
Keen EC (2015) A century of phage research: Bacteriophages and the shaping of modern biology. Bioessays 37:6–9
Klumpp J, Fouts DE, Sozhamannan S (2013) Bacteriophage functional genomics and its role in bacterial pathogen detection. Brief Funct Genomics 12:354–365
Jurczak-Kurek A, Gąsior T, Nejman-Faleńczyk B, Bloch S, Dydecka A et al (2016) Biodiversity of bacteriophages: morphological and biological properties of a large group of phages isolated from urban sewage. Sci Rep 6:34338
Hatfull GF (2015) Dark matter of the biosphere: the amazing world of bacteriophage diversity. Goodrum F, ed. J Virol 89:8107–8110
Ackermann HW (2001) Frequency of morphological phage descriptions in the year 2000. Arch Virol 146:843–857
Kutter E, Sulakvelidze A (2004) Bacteriophages: biology and applications. CRC Press, Boca Raton
Seeley ND, Primrose SB (1980) The effect of temperature on the ecology of aquatic bacteriophages. J Gen Virol 46:87–95
Klausa V, Piešinienė L, Staniulis J, Nivinskas R (2003) Abundance of T4-type bacteriophages in municipal wastewater and sewage. Ekologija 1:47–50
Leclerc H, Mossel DAA, Edberg SC, Struijk CB (2001) Advances in the bacteriology of the coliform group: their suitability as markers of microbial water safety. Annu Rev Microbiol 55:201–234
Yanagida M, Suzuki Y, Toda T (1984) Molecular organization of the head of bacteriophage T-even: underlying design principles. Adv Biophys 17:97–146
Wegrzyn G, Wegrzyn A (2005) Genetic switches during bacteriophage lambda development. Prog Nucleic Acid Res Mol Biol 79:1–48
Kaliniene L, Zajančkauskaitė A, Šimoliūnas E, Truncaitė L, Meškys R (2015) Low-temperature bacterial viruses VR—a small but diverse group of E. coli phages. Arch Virol 160:1367–1370
Šimoliūnas E, Kaliniene L, Stasilo M, Truncaitė L, Zajančkauskaitė A et al (2014) Isolation and characterization of vB_ArS-ArV2—first Arthrobacter sp. infecting bacteriophage with completely sequenced genome. PLoS One 9:e111230
Carlson K, Miller E (1994) Experiments in T4 genetics. In: Karam JD (ed) Bacteriophage T4. ASM Press, Washington DC, pp 419–483
Alva V, Nam SZ, Söding J, Lupas AN (2016) The MPI bioinformatics toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. doi:10.1093/nar/gkw348
Söding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33(suppl 2):W244–W248
Tamura K, Peterson D, Peterson N, Stecher G, Nei M et al (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32(suppl 2):W273–W279
Lopes A, Tavares P, Petit MA, Guérois R, Zinn-Justin S (2014) Automated identification of tailed bacteriophages and classification according to their neck organization. BMC Genomics 15:1027
Selick HE, Kreuzer KN, Alberts BM (1988) The bacteriophage T4 insertion/substitution vector system. A method for introducing site-specific mutations into the virus chromosome. J Biol Chem 263:11336–11347
Grenier F, Matteau D, Baby V, Rodrigue S (2014) Complete genome sequence of Escherichia coli BW25113. Genome Announc 2:e01038-14
Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2(2006):0008
Yoon SH, Han MJ, Jeong H, Lee CH, Xia XX, Lee DH, Shim JH, Lee SY, Oh TK, Kim JF (2012) Comparative multi-omics systems analysis of Escherichia coli strains B and K-12. Genome Biol 13:R37
Han MJ, Lee SY, Hong SH (2012) Comparative analysis of envelope proteomes in Escherichia coli B and K-12 strains. J Microbiol Biotechnol 22:470–478
Silhavy TJ, Kahne D, Walker S (2010) The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414
Müller-Loennies S, Lindner B, Brade H (2003) Structural analysis of oligosaccharides from lipopolysaccharide (LPS) of Escherichia coli K12 strain W3100 reveals a link between inner and outer core LPS biosynthesis. J Biol Chem 278:34090–34101
Jansson PE, Lindberg AA, Lindberg B, Wollin R (1981) Structural studies on the hexose region of the core in lipopolysaccharides from Enterobacteriaceae. Eur J Biochem 115:571–577
Bertozzi Silva J, Storms Z, Sauvageau D (2016) Host receptors for bacteriophage adsorption. FEMS Microbiol Lett 363(4):fnw002
Iyer LM, Koonin EV, Aravind L (2002) Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics 3:8
Warren RA (1980) Modified bases in bacteriophage DNAs. Annu Rev Microbiol 34:137–158
Young R (2014) Phage lysis: three steps, three choices, one outcome. J Microbiol 52:243–258
Rajaure M, Berry J, Kongari R, Cahill J, Young R (2015) Membrane fusion during phage lysis. Proc Natl Acad Sci USA 112:5497–5502
Rao VB, Feiss M (2008) The bacteriophage DNA packaging motor. Annu Rev Genet 42:647–681
Rao VB, Feiss M (2015) Mechanisms of DNA packaging by large double-stranded DNA viruses. Annu Rev Virol 2:351–378
Wietzorrek A, Schwarz H, Herrmann C, Braun V (2006) The genome of the novel phage Rtp, with a rosette-like tail tip, is homologous to the genome of phage T1. J Bacteriol 188:1419–1436
Casjens SR, Gilcrease EB (2009) Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol Biol 502:91–111
Stirm S, Bessler W, Fehmel F, Freund-Mölbert E (1971) Bacteriophage particles with endo-glycosidase activity. J Virol 8:343–346
Stummeyer K, Dickmanns A, Mühlenhoff M, Gerardy-Schahn R, Ficner R (2005) Crystal structure of the polysialic acid-degrading endosialidase of bacteriophage K1F. Nat Struct Mol Biol 12:90–96
Niu YD, McAllister TA, Nash JH, Kropinski AM, Stanford K (2014) Four Escherichia coli O157:H7 phages: a new bacteriophage genus and taxonomic classification of T1-like phages. PLoS One 9(6):e100426
Sharma M (2013) Lytic bacteriophages: potential interventions against enteric bacterial pathogens on produce. Bacteriophage 3:e25518
Painter JA, Hoekstra RM, Ayers T, Tauxe RV, Braden CR et al (2013) Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998–2008. Emerg Infect Dis 19:407–415
Ongeng D, Geeraerd AH, Springael D, Ryckeboer J, Muyanja C et al (2015) Fate of Escherichia coli O157:H7 and Salmonella enterica in the manure-amended soil-plant ecosystem of fresh vegetable crops: a review. Crit Rev Microbiol 41:273–294
Pérez Pulido R, Grande Burgos MJ, Gálvez A, Lucas López R (2016) Application of bacteriophages in post-harvest control of human pathogenic and food spoiling bacteria. Crit Rev Biotechnol 36:851–861
Acknowledgments
This work was supported by the Research Council of Lithuania (Project No. SIT-7/2015).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interests.
Human and animal rights statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Horst Neve.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kaliniene, L., Truncaitė, L., Šimoliūnas, E. et al. Molecular analysis of the low-temperature Escherichia coli phage vB_EcoS_NBD2. Arch Virol 163, 105–114 (2018). https://doi.org/10.1007/s00705-017-3589-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3589-5