Abstract
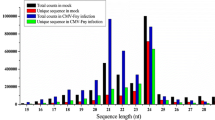
Cucumber green mottle mosaic virus (CGMMV) is a damaging pathogen that attacks crop plants belonging to the family Cucurbitaceae. Little is known about the regulatory role of microRNAs (miRNAs) in response to CGMMV infection. To identify CGMMV-responsive miRNAs, two sRNA libraries from mock-inoculated and CGMMV-infected watermelon leaves were constructed and sequenced using Solexa sequencing technology. In total, 471 previously known and 1,809 novel miRNAs were obtained, of which 377 known and 246 novel miRNAs were found to be differentially expressed during CGMMV infection. The target genes for the CGMMV-responsive known miRNAs are active in diverse biological processes, including cell wall modulation, plant hormone signaling, defense-related protein induction, primary and secondary metabolism, regulation of virus replication, and intracellular transport. The expression patterns of some CGMMV-responsive miRNAs and their corresponding targets were confirmed by RT-qPCR. One target gene for miR156a-5p was verified by 5’-RNA-ligase-mediated rapid amplification of cDNA ends (5’-RLM-RACE) analysis. The results of this study provide further insights into the miRNA-mediated regulatory network involved in the response to viral infection in watermelon and other cucurbit crops.






Similar content being viewed by others
References
Collins JK, Wu GY, Perkins-Veazie P, Spears K, Claypool PL, Baker RA, Clevidence BA (2007) Watermelon consumption increases plasma arginine concentrations in adults. Nutrition 23:261–266
Liu HW, Luo LX, Liang CQ, Jiang N, Liu PF, Li JQ (2015) High-throughput sequencing identifies novel and conserved cucumber (Cucumis sativus L.) microRNAs in response to cucumber green mottle mosaic virus infection. PLoS One 10:e0129002
Tesoriero LA, Chambers G, Srivastava M, Smith S, Conde B, Tran-Nguyen LTT (2016) First report of cucumber green mottle mosaic virus in Australia. Australas Plant Dis Notes 11:1
Tian T, Posis K, Maroon-Lango CJ, Mavrodieva V, Haymes S, Pitman TL, Falk BW (2014) First report of cucumber green mottle mosaic virus on melon in the United States. Plant Dis 98:1163
Liu HW, Luo LX, Li JQ, Liu PF, Chen XY, Hao JJ (2014) Pollen and seed transmission of cucumber green mottle mosaic virus in cucumber. Plant Pathol 63:72–77
Ainsworth GC (1935) Mosaic disease of the cucumber. Ann Appl Biol 22:55–67
Celix A, Luis-Arteaga M, Rodriguez-Cerezo E (1996) First report of cucumber green mottle mosaic tobamovirus infecting greenhouse-grown cucumber in Spain. Plant Dis 80:1303
Varveri C, Vassilakos N, Bem F (2002) Characterization and detection of cucumber green mottle mosaic virus in Greece. Phytoparasitica 30(5):93–501
Inouye T, Inouye N, Asatani M, Mitsuhata K (1967) Studies on cucumber green mottle mosaic virus in Japan. Ber Ohara Inst Landw Biol 14:49–69
Lee KW, Lee BC, Park HC, Lee YS (1990) Occurrence of cucumber green mottle mosaic virus disease of watermelon in Korea. Korean J Plant Pathol 6:250–255
Qin BX, Cai JH, Liu ZM, Chen YH, Zhu GN, Huang FX (2005) Preliminary identification of a cucumber green mottle mosaic virus infecting pumpkin. Plant Quar 4:198–200 (In Chinese)
Antignus Y, Pearlsman M, Ben-Yoseph R, Cohen S (1990) Occurrence of a variant of cucumber green mottle mosaic virus in Israel. Phytoparasitica 18:50–56
Al-Shahwan IM, Abdalla OA (1992) A strain of cucumber green mottle mosaic virus (CGMMV) from bottlegourd in Saudi Arabia. J Phytopathol 134:152–156
Ling K, Li R, Zhang W (2014) First report of cucumber green mottle mosaic virus infecting greenhouse cucumber in Canada. Plant Dis 98:701
Wu HJ, Qin BX, Chen HY, Peng B, Cai JH, Gu QS (2011) The rate of seed contamination and transmission of cucumber green mottle mosaic virus in watermelon and melon. Sci Agric Sin 44:1527–1532
Li J, Gu Q (2015) Research progress on transmission modes of cucumber green mottle mosaic virus. China Veg 1:13–18 (in Chinese)
Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC (2003) P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev Cell 4:205–217
Khraiwesh B, Zhu JK, Zhu J (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta 1819:137–148
Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25:2383–2399
Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J (2012) MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA 109:1790–1795
Du P, Wu J, Zhang J, Zhao S, Zheng H, Gao G, Wei LP, Li Y (2011) Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog 7:e1002176
Yin X, Wang J, Cheng H, Wang X, Yu D (2013) Detection and evolutionary analysis of soybean miRNAs responsive to soybean mosaic virus. Planta 237:1213–1225
Feng J, Liu S, Wang M, Lang Q, Jin C (2014) Identification of microRNAs and their targets in tomato infected with cucumber mosaic virus based on deep sequencing. Planta 240:1335–1352
Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967
Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A (2005) Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res 33:121–124
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2011) GenBank. Nucleic Acids Res 39:D32–D37
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36:154–158
Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao XF, Carrington JC, Chen X, Green PJ, Griffiths-Jones S, Jacobsen SE, Mallory AC, Martienssen RA, Scott Poethig R, Qi Y, Vaucheret H, Voinnet O, Watanabe Y, Weigel D, Zhu J (2008) Criteria for annotation of plant microRNAs. Plant Cell 20:3186–3190
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Eldem V, Akcay UC, Ozhuner E, Bakir Y, Uranbey S, Unver T (2012) Genome-wide identification of miRNAs responsive to drought in peach (Prunus persica) by high-throughput deep sequencing. PLoS One 7:e50298
Li BS, Qin YR, Duan H, Yin WL, Xia XL (2011) Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J Exp Bot 62:3765–3779
Allen E, Xie Z, Gustafson AM, Carrington JC (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121:207–221
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8:517–527
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:182–185
Shi R, Chiang VL (2005) Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39:519–525
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408
Ramesh SV, Ratnaparkhe MB, Kumawat G, Gupta GK, Husain SM (2014) Plant miRNAome and antiviral resistance: a retrospective view and prospective challenges. Virus Genes 48:1–14
He X, Fang Y, Feng L, Guo H (2008) Characterization of conserved novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett 582:2445–2452
Yang J, Zheng SL, Zhang HM, Liu XY, Li J, Li JM, Chen JP (2014) Analysis of small RNAs derived from Chinese wheat mosaic virus. Arch Virol 159:3077–3082
Li Y, Deng C, Shang Q, Zhao X, Liu X, Zhou Q (2016) Characterization of siRNAs derived from cucumber green mottle mosaic virus in infected cucumber plants. Arch Virol 161:455–458
Ding SW, Voinnet O (2007) Antiviral immunity directed by small RNAs. Cell 130:413–426
Wang XB, Wu QF, Ito T, Cillo F, Li WX, Chen X, Yu J, Ding S (2010) RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc Natl Acad Sci USA 107:484–489
Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One 2:e219
Bazzini AA, Hopp HE, Beachy RN, Asurmendi S (2007) Infection and co-accumulation of tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development. Proc Natl Acad Sci USA 104:12157–12162
van Loon LC, Rep M, Pieterse CM (2006) Significance of inducible defence-related proteins in infected plants. Annu Rev Phytopathol 44:35–162
Lu S, Li Q, Wei H, Chang MJ, Tunlaya AS, Kim H, Liu J, Song J, Sun Y, Yuan L, Yeh T, Peszlen I, Ralph J, Sederoff SR, Chiang VL (2013) Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc Natl Acad Sci USA 110:10848–10853
Rhee Y, Tzfira T, Chen M, Waigmann E, Citovsky V (2001) Cell-to-cell movement of Tobacco mosaic virus: enigmas and explanations. Mol Plant Pathol 1:33–39
Kima W, Kima J, Koe J, Kimd J, Hana K (2013) Transcription factor MYB46 is an obligate component of the transcriptional regulatory complex for functional expression of secondary wall-associated cellulose synthases in Arabidopsis thaliana. J Plant Physiol 170:1374–1378
Adie BA, Perez-Perez J, Perez-Perez MM, Godoy M, Sanchez-Serrano JJ, Schmelz EA, Solano R (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defences in Arabidopsis. Plant Cell 19:1665–1681
Padmanabhan MS, Kramer SR, Wang X, Culver JN (2008) Tobacco mosaic virus replicase-auxin/indole acetic acid protein interactions: reprogramming the auxin response pathway to enhance virus infection. J Virol 82:2477–2485
Navarro L, Bari R, Achard P, Lison P, Nemri A, Harberd NP, Jones JD (2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 18:650–655
McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139:949–959
Siemens J, Keller I, Sarx J, Kunz S, Schuller A, Nagel W, Schmulling T, Parniske M, Ludwig-Muller J (2006) Transcriptome analysis of Arabidopsis clubroots indicate a key role for cytokinins in disease development. Mol Plant Microbe Interact 19:480–494
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444:323–329
Robaglia C, Caranta C (2006) Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci 11:40–45
Ray S, Yumak H, Domashevskiy A, Khan MA, Gallie DR, Goss DJ (2006) Tobacco etch virus mRNA preferentially binds wheat germ eukaryotic initiation factor (eIF) 4G rather than eIFiso4G. J Biol Chem 281:35826–35834
Ayme V, Petit-Pierre J, Souche S, Palloix A, Moury B (2007) Molecular dissection of the potato virus Y VPg virulence factor reveals complex adaptations to the pvr2 resistance allelic series in pepper. J Gen Virol 88:1594–1601
Albar L, Bangratz-Reyser M, Hebrard E, Ndjiondjop MN, Jones M, Ghesquiere A (2006) Mutations in the eIF(iso)4G translation initiation factor confer high resistance of rice to Rice yellow mottle virus. Plant J 47:417–426
Boisnard A, Albar L, Thiéméle D, Rondeau M, Ghesquière A (2007) Evaluation of genes from eIF4E and eIF4G multigenic families as potential candidates for partial resistance QTLs to Rice yellow mottle virus in rice. Theor Appl Genet 116:53–62
Santiago R, Malvar RA (2010) Role of dehydrodiferulates in maize resistance to pests and diseases. Int J Mol Sci 11:691–703
Sampietro DA, Fauguel CM, Vattuone MA, Presello DA, Catalán CAN (2013) Phenylpropanoids from maize pericarp: resistance factors to kernel infection and fumonisin accumulation by Fusarium verticillioides. Eur J Plant Pathol 135:105–113
Wang J, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138(4):738–749
Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23:1512–1522
Mao H, Yu L, Li Z, Yan Y, Han R, Ma M (2016) Genome-wide analysis of the SPL family transcription factors and their responses to abiotic stresses in maize. Plant Gene 6:1–12
Alizadeh F, Abdullah SNS, Chong P, Selamat AB (2014) Expression analysis of fatty acid biosynthetic pathway genes during interactions of oil palm (Elaeis guineensis Jacq.) with the pathogenic Ganoderma boninense and symbiotic Trichoderma harzianum fungal organisms. Plant Mol Biol Rep 32:70–81
Nakaune R, Hamamoto H, Imada J, Akutsu K, Hibi T (2002) A novel ABC transporter gene, PMR5, is involved in multidrug resistance in the phytopathogenic fungus Penicillium digitatum. Mol Genet Genom 267:179–185
Wang Y, Chai C, Valliyodan B, Maupin C, Annen B, Nguyen HT (2015) Genome-wide analysis and expression profiling of the PIN auxin transporter gene family in soybean (Glycine max). BMC Genom 16:951
Howarth JR, Fourcroy P, Davidian J, Smith FW, Hawkesford MJ (2003) Cloning of two contrasting high-affinity sulfate transporters from tomato induced by low sulfate and infection by the vascular pathogen Verticillium dahliae. Planta 218:58–64
Joko T, Hirata H, Tsuyumu S (2007) A sugar transporter (MfsX) is also required by Dickeya dadantii 3937 for in planta fitness. J Gen Plant Pathol 73:274–280
Eybishtz A, Peretz Y, Sade D, Gorovits R, Czosnek H (2010) Tomato yellow leaf curl virus infection of a resistant tomato line with a silenced sucrose transporter gene LeHT1 results in inhibition of growth, enhanced virus spread, and necrosis. Planta 231:537–548
Acknowledgements
This work was funded by grants from the China Postdoctoral Science Foundation (2016M601973), the National Natural Science Foundation of China (31572145).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Human and animal rights statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2017_3401_MOESM2_ESM.doc
Supplementary material 2 Table S1 Primers used for quantification and validation of selected watermelon CGMMV responsive miRNAs and their targets (DOC 35 kb)
705_2017_3401_MOESM4_ESM.xlsx
Supplementary material 4 Table S3 Novel miRNA precursor candidates identified from mock and CGMMV libraries (XLSX 482 kb)
Rights and permissions
About this article
Cite this article
Sun, Y., Niu, X. & Fan, M. Genome-wide identification of cucumber green mottle mosaic virus-responsive microRNAs in watermelon. Arch Virol 162, 2591–2602 (2017). https://doi.org/10.1007/s00705-017-3401-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3401-6