Abstract
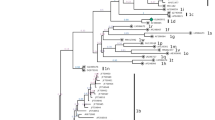
Bovine viral diarrhea virus 1 (BVDV-1) belongs to the genus Pestivirus within the family Flaviviridae. Based on the 5’ untranslated region (UTR) sequence, BVDV-1 can be divided into at least 17 subtypes (1a though 1q). BVDV-1i is an uncommon subtype that has been reported in the United Kingdom and Uruguay. Here, we report the complete genome sequence of the first subtype 1i BVDV-1 (strain ACM/BR/2016) isolated from cattle in southern Brazil. The genome is 12,231 nt in length and contains a single ORF that encodes a polyprotein of 3,896 amino acids, flanked by 5’ and 3’UTRs of 325 and 220 nt, respectively. Phylogenetic inferences based on the whole genome, the 5’UTR, and the Npro region showed that strain ACM/BR/2016 is closely related to previously characterized BVDV-1i members. Its 5’UTR shares the highest nucleotide identity (90.5%) with BVDV-1i strains from United Kingdom, and its Npro is most closely related to that of a Uruguayan strain (90.6%). To the best of our knowledge, this is the first BVDV-1i strain from which the whole genome has been completely sequenced and characterized. The complete genome of a BVDV-1i will help future studies on pestivirus evolution and heterogeneity.

Similar content being viewed by others
References
Simmonds P, Becher P, Collet MS et al (2011) Flaviviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus Taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, pp 1003–1020
Avalos-Ramirez R, Orlich M, Thiel HJ, Becher P (2001) Evidence for the presence of two novel pestivirus species. Virology 286:456–465. doi:10.1006/viro.2001.1001
Vilcek S, Ridpath JF, Van Campen H et al (2005) Characterization of a novel pestivirus originating from a pronghorn antelope. Virus Res 108:187–193. doi:10.1016/j.virusres.2004.09.010
Kirkland PD, Frost MJ, Finlaison DS et al (2007) Identification of a novel virus in pigs—Bungowannah virus: a possible new species of pestivirus. Virus Res 129:26–34. doi:10.1016/j.virusres.2007.05.002
Schirrmeier H, Strebelow G, Depner K et al (2004) Genetic and antigenic characterization of an atypical pestivirus isolate, a putative member of a novel pestivirus species. J Gen Virol 85:3647–3652. doi:10.1099/vir.0.80238-0
Houe H (2003) Economic impact of BVDV infection in dairies. Biologicals 31:137–143. doi:10.1016/S1045-1056(03)00030-7
Vilcek S, Paton DJ, Durkovic B et al (2001) Bovine viral diarrhoea virus genotype 1 can be separated into at least eleven genetic groups. Arch Virol 146:99–115
Deng Y, Shan T-L, Tong W et al (2014) Genomic characterization of a bovine viral diarrhea virus 1 isolate from swine. Arch Virol. doi:10.1007/s00705-014-2064-9
Weber MN, Silveira S, Machado G et al (2014) High frequency of bovine viral diarrhea virus type 2 in Southern Brazil. Virus Res 191:117–124. doi:10.1016/j.virusres.2014.07.035
Giammarioli M, Ridpath JF, Rossi E et al (2015) Genetic detection and characterization of emerging HoBi-like viruses in archival foetal bovine serum batches. Biologicals 43:220–224. doi:10.1016/j.biologicals.2015.05.009
Ridpath JF, Lovell G, Neill JD et al (2011) Change in predominance of Bovine viral diarrhea virus subgenotypes among samples submitted to a diagnostic laboratory over a 20-year time span. J Vet Diagn Investig 23:185–193. doi:10.1177/104063871102300201
Booth RE, Thomas CJ, El-Attar LMR et al (2013) A phylogenetic analysis of bovine viral diarrhoea virus (BVDV) isolates from six different regions of the UK and links to animal movement data. Vet Res 44:43. doi:10.1186/1297-9716-44-43
Strong R, Errington J, Cook R et al (2013) Increased phylogenetic diversity of bovine viral diarrhoea virus type 1 isolates in England and Wales since 2001. Vet Microbiol 162:315–320. doi:10.1016/j.vetmic.2012.09.006
Maya L, Puentes R, Reolón E et al (2016) Molecular diversity of bovine viral diarrhea virus in uruguay. Arch Virol 161:529–535. doi:10.1007/s00705-015-2688-4
Vilcek S, Herring AJ, Herring JA et al (1994) Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch Virol 136:309–323
Bankevich A, Nurk S, Antipov D et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi:10.1089/cmb.2012.0021
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi:10.1093/nar/gkf436
Tamura K, Stecher G, Peterson D et al (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
Acknowledgements
The authors thank Setor de Patologia Veterinária (Faculdade de Veterinária, Universidade Federal do Rio Grande do Sul) for providing isolates, and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and PROPESQ/UFRGS for supporting this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors whose names are listed below certify that they have no affiliations with or involvement in any organization or entity with any financial interest (such as honoraria, educational grants, participation in speakers’ bureaus, membership employment, consultancies, stock ownership, or other equity interest, and expert testimony or patent-licensing arrangements), or non-financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript. We have no conflict of interest.
Statement on the welfare of animals
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Mósena, A.C.S., Weber, M.N., Cibulski, S.P. et al. Genomic characterization of a bovine viral diarrhea virus subtype 1i in Brazil. Arch Virol 162, 1119–1123 (2017). https://doi.org/10.1007/s00705-016-3199-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-016-3199-7