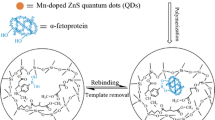
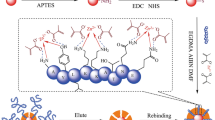
Abstract
Molecularly imprinted polymer (MIP) nanofilms for alpha-fetoprotein (AFP) and the receptor binding domain (RBD) of the spike protein of SARS-CoV-2 using either a peptide (epitope-MIP) or the whole protein (protein-MIP) as the template were prepared by electropolymerization of scopoletin. Conducting atomic force microscopy revealed after template removal and electrochemical deposition of gold a larger surface density of imprinted cavities for the epitope-imprinted polymers than when using the whole protein as template. However, comparable affinities towards the respective target protein (AFP and RBD) were obtained for both types of MIPs as expressed by the KD values in the lower nanomolar range. On the other hand, while the cross reactivity of both protein-MIPs towards human serum albumin (HSA) amounts to around 50% in the saturation region, the nonspecific binding to the respective epitope-MIPs is as low as that for the non-imprinted polymer (NIP). This effect might be caused by the different sizes of the imprinted cavities. Thus, in addition to the lower costs the reduced nonspecific binding is an advantage of epitope-imprinted polymers for the recognition of proteins.
Graphical Abstract






Similar content being viewed by others
References
Polyakov MV (1931) Adsorption properties and structure of silica gel. Zhur Fiz Khim 2:799–895
Wulff G, Sarhan A (1972) Über die Anwendung von enzymanalog gebauten Polymeren zur Racemattrennung. Angew Chemie 84:364–364. https://doi.org/10.1002/ange.19720840838
Arshady R, Mosbach K (1981) Synthesis of substrate-selective polymers by host-guest polymerization. Die Makromol Chemie 182:687–692. https://doi.org/10.1002/MACP.1981.021820240
Birnbaumer GM, Lieberzeit PA, Richter L et al (2009) Detection of viruses with molecularly imprinted polymers integrated on a microfluidic biochip using contact-less dielectric microsensors. Lab Chip 9:3549–3556. https://doi.org/10.1039/B914738A
Amorim MS, Sales MGF, Frasco MF (2022) Recent advances in virus imprinted polymers. Biosens Bioelectron X 10:100131. https://doi.org/10.1016/J.BIOSX.2022.100131
Rachkov A, Minoura N (2000) Recognition of oxytocin and oxytocin-related peptides in aqueous media using a molecularly imprinted polymer synthesized by the epitope approach. J Chromatogr A 889:111–118
Rachkov A, Minoura N (2001) Towards molecularly imprinted polymers selective to peptides and proteins. The epitope approach. Biochim Biophys Acta - Protein Struct Mol Enzymol 1544:255–266. https://doi.org/10.1016/S0167-4838(00)00226-0
Yoshimatsu K, Yamazaki T, Hoshino Y et al (2014) Epitope discovery for a synthetic polymer nanoparticle: a new strategy for developing a peptide tag. J Am Chem Soc 136:1194–1197. https://doi.org/10.1021/ja410817p
Dechtrirat D, Jetzschmann KJ, Stöcklein WFM et al (2012) Protein rebinding to a surface-confined imprint. Adv Funct Mater 22:5231–5237. https://doi.org/10.1002/adfm.201201328
Moczko E, Guerreiro A, Cáceres C et al (2019) Epitope approach in molecular imprinting of antibodies. J Chromatogr B 1124:1–6. https://doi.org/10.1016/j.jchromb.2019.05.024
Nishino H, Huang CS, Shea KJ (2006) Selective protein capture by epitope imprinting. Angew Chemie - Int Ed 45:2392–2396. https://doi.org/10.1002/anie.200503760
Singh M, Gupta N, Raghuwanshi R (2017) Epitope imprinting approach to monitor diseases. J Mol Genet Med 11:1–6. https://doi.org/10.4172/1747-0862.1000270
Bosserdt M, Gajovic-Eichelman N, Scheller FW (2013) Modulation of direct electron transfer of cytochrome c by use of a molecularly imprinted thin film. Anal Bioanal Chem 405:6437–6444. https://doi.org/10.1007/s00216-013-7009-8
Wright LM, Kreikemeier JT, Fimmel CJ (2007) A concise review of serum markers for hepatocellular cancer. Cancer Detect Prev 31:35–44. https://doi.org/10.1016/J.CDP.2006.11.003
Yuen MF, Lai CL (2005) Serological markers of liver cancer. Best Pract Res Clin Gastroenterol 19:91–99. https://doi.org/10.1016/J.BPG.2004.10.003
Liu Z, Xiao X, Wei X et al (2020) Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol 92:595–601. https://doi.org/10.1002/JMV.25726
Zhang X, Waffo AT, Yarman A et al (2022) How an ACE2 mimicking epitope-MIP nanofilm recognizes template-related peptides and the receptor binding domain of SARS-CoV-2. Nanoscale 14:18106–18114. https://doi.org/10.1039/D2NR03898F
Settipani J, Karim K, Chauvin A et al (2018) Theoretical aspects of peptide imprinting: screening of MIP (virtual) binding sites for their interactions with amino acids, di- and tripeptides. J Chinese Adv Mater Soc 6:301–310. https://doi.org/10.1080/22243682.2018.1467279
Bognár Z, Supala E, Yarman A et al (2022) Peptide epitope-imprinted polymer microarrays for selective protein recognition. Application for SARS-CoV-2 RBD protein. Chem Sci 13:1263–1269. https://doi.org/10.1039/D1SC04502D
Neusser G, Eppler S, Bowen J et al (2017) FIB and MIP: understanding nanoscale porosity in molecularly imprinted polymers via 3D FIB/SEM tomography. Nanoscale 9:14327–14334. https://doi.org/10.1039/C7NR05725C
Kondo T, Udagawa I, Aikawa T et al (2016) Enhanced sensitivity for electrochemical detection using screen-printed diamond electrodes via the random microelectrode array effect. Anal Chem 88:1753–1759. https://doi.org/10.1021/ACS.ANALCHEM.5B03986/ASSET/IMAGES/LARGE/AC-2015-03986V_0008.JPEG
Zhang X, Yarman A, Erdossy J et al (2018) Electrosynthesized MIPs for transferrin: plastibodies or nano-filters? Biosens Bioelectron 105:29–35. https://doi.org/10.1016/J.BIOS.2018.01.011
Taheri N, Khoshsafar H, Ghanei M et al (2022) Dual-template rectangular nanotube molecularly imprinted polypyrrole for label-free impedimetric sensing of AFP and CEA as lung cancer biomarkers. Talanta 239:123146. https://doi.org/10.1016/J.TALANTA.2021.123146
Cieplak M, Szwabinska K, Sosnowska M et al (2015) Selective electrochemical sensing of human serum albumin by semi-covalent molecular imprinting. Biosens Bioelectron 74:960–966. https://doi.org/10.1016/j.bios.2015.07.061
Bosserdt M, Erdőssy J, Lautner G et al (2015) Microelectrospotting as a new method for electrosynthesis of surface-imprinted polymer microarrays for protein recognition. Biosens Bioelectron 73:123–129. https://doi.org/10.1016/j.bios.2015.05.049
Cai D, Ren L, Zhao H et al (2010) A molecular-imprint nanosensor for ultrasensitive detection of proteins. Nat Nanotechnol 5:597–601. https://doi.org/10.1038/nnano.2010.114
Karimian N, Vagin M, Zavar MHA et al (2013) An ultrasensitive molecularly-imprinted human cardiac troponin sensor. Biosens Bioelectron 50:492–498. https://doi.org/10.1016/j.bios.2013.07.013
Yarman A (2018) Electrosynthesized molecularly imprinted polymer for laccase using the inactivated enzyme as the target. Bull Korean Chem Soc 39:483–488. https://doi.org/10.1002/bkcs.11413
Drobysh M, Ramanaviciene A, Viter R et al (2022) Biosensors for the determination of SARS-CoV-2 virus and diagnosis of COVID-19 infection. Int J Mol Sci 23:666. https://doi.org/10.3390/IJMS23020666
Ayankojo AG, Boroznjak R, Reut J et al (2022) Molecularly imprinted polymer based electrochemical sensor for quantitative detection of SARS-CoV-2 spike protein. Sensors Actuators B Chem 353:131160. https://doi.org/10.1016/J.SNB.2021.131160
Palladino P, Minunni M, Scarano S (2018) Cardiac troponin T capture and detection in real-time via epitope-imprinted polymer and optical biosensing. Biosens Bioelectron 106:93–98. https://doi.org/10.1016/J.BIOS.2018.01.068
Zhao C, Ma X, Li J (2017) An insulin molecularly imprinted electrochemical sensor based on epitope imprinting. Chinese J Anal Chem 45:1360–1366. https://doi.org/10.1016/S1872-2040(17)61039-9
Cheng Z, Zhang L, Li Y (2004) Synthesis of an enzyme-like imprinted polymer with the substrate as the template, and its catalytic properties under aqueous conditions. Chem – A Eur J 10:3555–3561. https://doi.org/10.1002/CHEM.200305370
Li S, Yang K, Liu J et al (2015) Surface-imprinted nanoparticles prepared with a His-tag-anchored epitope as the template. Anal Chem 87:4617–4620. https://doi.org/10.1021/ac5047246
Li S, Yang K, Zhao B et al (2016) Epitope imprinting enhanced IMAC (EI-IMAC) for highly selective purification of His-tagged protein. J Mater Chem B 4:1960–1967. https://doi.org/10.1039/C5TB02505B
Bognár Z, Kozma J, Kovács N, Gyurcsányi RE (2023) Novel functional monomer for the electrochemical synthesis of highly affine epitope-imprinted polymers. Electroanalysis e202300025. https://doi.org/10.1002/ELAN.202300025
Yang K, Li S, Liu J et al (2016) Multiepitope templates imprinted particles for the simultaneous capture of various target proteins. Anal Chem 88:5621–5625. https://doi.org/10.1021/acs.analchem.6b01247
Erdőssy J, Horváth V, Yarman A et al (2016) Electrosynthesized molecularly imprinted polymers for protein recognition. TrAC Trends Anal Chem 79:179–190. https://doi.org/10.1016/J.TRAC.2015.12.018
Zhang X, Caserta G, Yarman A et al (2021) “Out of pocket” protein binding—a dilemma of epitope imprinted polymers revealed for human hemoglobin. Chemosensors 9:128. https://doi.org/10.3390/CHEMOSENSORS9060128
Amouzadeh Tabrizi M, Fernández-Blázquez JP, Medina DM, Acedo P (2022) An ultrasensitive molecularly imprinted polymer-based electrochemical sensor for the determination of SARS-CoV-2-RBD by using macroporous gold screen-printed electrode. Biosens Bioelectron 196:113729. https://doi.org/10.1016/J.BIOS.2021.113729
Parisi OI, Francomano F, Dattilo M et al (2022) The evolution of molecular recognition: from antibodies to molecularly imprinted polymers (MIPs) as artificial counterpart. J Funct Biomater 13:12. https://doi.org/10.3390/JFB13010012
Raziq A, Kidakova A, Boroznjak R et al (2021) Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens Bioelectron 178:113029. https://doi.org/10.1016/j.bios.2021.113029
Zang Y, Zhang Y, Wei R et al (2023) Difunctional molecularly imprinted polymers and heterostructured CdS nanoparticle-sensitized ZnO nanorod arrays for antibody-free photoelectrochemical alpha-fetoprotein sensor. J Electroanal Chem 944:117631. https://doi.org/10.1016/J.JELECHEM.2023.117631
Shen X, Ma Y, Zeng Q et al (2016) Molecularly imprinted electrochemical sensor for advanced diagnosis of alpha-fetoprotein. Anal Methods 8:7361–7368. https://doi.org/10.1039/c6ay01922f
Wu Y, Wang Y, Wang X et al (2019) Electrochemical sensing of α-fetoprotein based on molecularly imprinted polymerized ionic liquid film on a gold nanoparticle modified electrode surface. Sensors (Switzerland) 19:3218. https://doi.org/10.3390/s19143218
Lai Y, Zhang C, Deng Y et al (2019) A novel α-fetoprotein-MIP immunosensor based on AuNPs/PTh modified glass carbon electrode. Chinese Chem Lett 30:160–162. https://doi.org/10.1016/J.CCLET.2018.07.011
Scheller F, Renneberg R (1982) Electrochemical studies of proteins. In: Dryhurst G, Kadish KM, Scheller F, Renneberg R (eds) Biological electrochemistry, 1st edn. Academic Press, London, UK, pp 398–521
Acknowledgements
The authors would like to thank the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy—EXC 2008–390540038 (Unifying Systems in Catalysis—UniSysCat) and the German Ministry of Education and Research (BMBF, 01DH20018). This work was partially supported by the Ministry of Innovation and Technology of Hungary from the National Research, Development and Innovation Fund under the TKP2021 funding scheme, project no. BME-EGA-02.
Author information
Authors and Affiliations
Contributions
Conceptualization, R.E.G. and F.W.S.; methodology and formal analysis, X.Z., A.Y., N.K., Z.B., and R.E.G.; investigation, X.Z., N.K., and Z.B.; writing—original draft preparation, X.Z., R.E.G., and F.W.S.; writing—review and editing, R.E.G. and F.W.S.; visualization, X.Z. Z.B., and N.K.; supervision, A.Y., R.E.G., and F.W.S.; project administration, F.W.S.; funding acquisition, R.E.G., F.F.B., and F.W.S. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
The paper is dedicated to both pioneers of MIP development Günther Wulff and Klaus Mosbach.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, X., Yarman, A., Kovács, N. et al. Specific features of epitope-MIPs and whole-protein MIPs as illustrated for AFP and RBD of SARS-CoV-2. Microchim Acta 191, 242 (2024). https://doi.org/10.1007/s00604-024-06325-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-024-06325-0