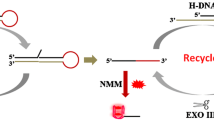
Abstract
A simple, rapid, and highly efficient fluorescent detection technique without PCR through dual-probe ligation with the genetic capture of magnetic beads and reported probe was developed for determination of epidermal growth factor receptor (EGFR) gene exon 19 deletions. The EGFR exon 19 deletion mutation makes up 48% of all mutations associated with anti-tyrosine kinase inhibition sensitivity, and thus, the EGFR nucleotide variant is very important in clinical diagnosis. In this approach, the dual-probe ligation was designed to target exon 19 deletion. The magnetic genetic captured system was then applied to capture the successful dual-probe ligation. Thereafter, a reporter probe which is coupled with 6-fluorescein amidite (6-FAM) was introduced to hybridize with dual-probe ligation product on the surface of streptavidin magnetic beads, and finally, the supernatant was taken for fluorescence measurements for distinguishing mutant types from wild types. After optimization (the RSD of the fluorescent intensity was less than 4.5% (n = 3) under the optimal condition), 20 blind DNA samples from the population were analyzed by this technique and further confirmed by direct sequencing. The results of our assay matched to those from direct sequencing data, evidencing that the developed method is accurate and successful. These 20 blind DNA samples were diagnosed as wild and then spiked with different percentages of the mutant gene to quantify the ratio of the wild and mutant genes. This strategy was also successfully applied to quantify the ratio of the wild and mutant genes with good linearity at the λex/λem of 480 nm/520 nm (r = 0.996), and the limit of detection reached 1.0% mutant type. This simple fluorescent detection of nucleotide variants shows its potential to be considered a tool in biological and clinical diagnosis. Importantly, this strategy offers a universal detection capability for any kind of mutation (point, deletion, insertion, or substitution) in a gene of interest.
Graphical abstract








Similar content being viewed by others
Abbreviations
- EGFR:
-
Epidermal growth factor receptor
- 6-FAM:
-
6-Fluorescein amidite
- DNA:
-
Deoxyribonucleic acid
- PCR:
-
Polymerase chain reaction
- NSCLC:
-
Non-small cell lung cancer
- TKI:
-
Tyrosine kinase inhibitors
- PNA-LNA:
-
Peptide nucleic acid-locked nucleic acid
- ARMS:
-
Amplification refractory mutation system
- HPLC:
-
High-performance liquid chromatography
- RFLP:
-
Restriction fragment length polymorphism
- RCA:
-
Rolling circle amplification
References
Shendure J, Findlay GM, Snyder MW (2019) Genomic medicine-progress, pitfalls, and promise. Cell 177:45–57. https://doi.org/10.1016/j.cell.2019.02.003
Wise AL, Manolio TA, Mensah GA, Peterson JF, Roden DM, Tamburro C, Williams MS, Green ED (2019) Genomic medicine for undiagnosed diseases. Lancet 394:533–540. https://doi.org/10.1016/S0140-6736(19)31274-7
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A (2015) Global cancer statistics, 2012. CA Cancer J Clin 65:87–108. https://doi.org/10.3322/caac.21262
Herbst RS, Morgensztern D, Boshoff C (2018) The biology and management of non-small cell lung cancer. Nature 553:446–454. https://doi.org/10.1038/nature25183
Bae JH, Jo SM, Kim HS (2015) Comprehensive detection of diverse exon 19 deletion mutations of EGFR in lung cancer by a single probe set. Biosens Bioelectron 74:849–855. https://doi.org/10.1016/j.bios.2015.07.043
Ahlawat P, Phutela K, Bal A, Singh N, Sharma S (2022) Therapeutic potential of human serum albumin nanoparticles encapsulated actinonin in murine model of lung adenocarcinoma. Drug Deliv 29:2403–2413. https://doi.org/10.1080/10717544.2022.2067600
Xu XW, Weng XH, Wang CL, Lin WW, Liu AL, Chen W, Lin XH (2016) Detection EGFR exon 19 status of lung cancer patients by DNA electrochemical biosensor. Biosens Bioelectron 80:411–417. https://doi.org/10.1016/j.bios.2016.02.009
Greenhalgh J, Boland A, Bates V, Vecchio F, Dundar Y, Chaplin M, Green JA (2021) First-line treatment of advanced epidermal growth factor receptor (EGFR) mutation positive non-squamous non-small cell lung cancer. Cochrane Database Syst Rev 3:CD010383. https://doi.org/10.1002/14651858.CD010383.pub3
Roengvoraphoj M, Tsongalis GJ, Dragnev KH, Rigas JR (2013) Epidermal growth factor receptor tyrosine kinase inhibitors as initial therapy for non-small cell lung cancer: focus on epidermal growth factor receptor mutation testing and mutation-positive patients. Cancer Treat Rev 39:839–850. https://doi.org/10.1016/j.ctrv.2013.05.001
Ren KH, Qin WW, Wang Y, Peng JC, Hu WX (2022) Detection of an EML4-ALK fusion mutation secondary to epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) therapy for lung cancer: a case report. Ann Palliat Med 11:2503–2509. https://doi.org/10.21037/apm-22-744
Salmani H, Yahyaei M, Raoofian R, Irani S, Salahshouri Far I, Akrami SM (2022) Evaluation of epidermal growth factor receptor gene mutations in an Iranian population with non-small cell lung carcinoma. Iran J Public Health 51:450–459. https://doi.org/10.18502/ijph.v51i2.8698
Almeida SB, Spencer AS, Santos CLD, Fernandes G, Simões P, Silva S, Domingues TD, Honório M (2022) Switch to EGFR-TKI after upfront platinum doublet induction therapy in non-small cell lung cancer (NSCLC) patients with EGFR (Epidermal Growth Factor Receptor) mutation: a multicentre retrospective study. Cancer Treat Res Commun 31:100526. https://doi.org/10.1016/j.ctarc.2022.100526
Mitsudomi T, Yatabe Y (2010) Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J 277:301–308. https://doi.org/10.1111/j.1742-4658.2009.07448.x
Keedy VL, Temin S, Somerfield MR, Beasley MB, Johnson DH, McShane LM, Milton DT, Strawn JR, Wakelee HA, Giaccone G (2011) American society of clinical oncology provisional clinical opinion: epidermal growth factor receptor (EGFR) mutation testing for patients with advanced non-small-cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol 29:2121–2127. https://doi.org/10.1200/JCO.2010.31.8923
Chabot-Richards D, Buehler K, Vasef M (2015) Detection of EGFR exon 19 E746-A750 deletion and EGFR exon 21 point mutations in lung adenocarcinoma by immunohistochemistry: a comparative study to EGFR exons 19 and 21 mutations analysis using PCR followed by high-resolution melting and pyrosequencing. J Histotechnol 38(2046023615Y):000. https://doi.org/10.1179/2046023615Y.0000000002
Ellison G, Zhu G, Moulis A, Dearden S, Speake G, McCormack R (2013) EGFR mutation testing in lung cancer: a review of available methods and their use for analysis of tumour tissue and cytology samples. J Clin Pathol 66:79–89. https://doi.org/10.1136/jclinpath-2012-201194
Morise M, Taniguchi H, Saka H, Shindoh J, Suzuki R, Kojima E, Hase T, Ando M, Kondo M, Saito H, Hasegawa Y (2014) Phase II study of erlotinib for previously treated patients with EGFR wild-type non-small-cell lung cancer, following EGFR mutation status reevaluation with the scorpion amplified refractory mutation system. Mol Clin Oncol 2:991–996. https://doi.org/10.3892/mco.2014.354
Gao W, He J, Jin SD, Xu J, Yu TF, Wang W, Zhu Q, Dai H, Wu H, Liu YQ, Shu YQ, Guo RH (2019) Association of initial epidermal growth factor receptor tyrosine kinase inhibitors treatment and EGFR exon 19 deletion with frequency of the T790M mutation in non-small cell lung cancer patients after resistance to first-line epidermal growth factor receptor tyrosine kinase inhibitors. Onco Targets Ther 12:9495–9504. https://doi.org/10.2147/OTT.S220383
Zhang Y, Huang X, Niu R, Li C, Pang J, Liu P, Adachi H, Kawase A, Yamaguchi F, Du Y (2022) Association between EGFR gene mutant protein expression and T790M mutation after first-generation EGFR-TKI treatment resistance: a retrospective, single-arm clinical study. Ann Transl Med 10:935. https://doi.org/10.21037/atm-22-3850
Guo K, Shao C, Han L, Liu H, Ma Z, Yang Y, Feng Y, Pan M, Santarpia M, Carmo-Fonseca M, Silveira C, Lee KY, Han J, Li X, Yan X (2021) Detection of epidermal growth factor receptor (EGFR) mutations from preoperative circulating tumor DNA (ctDNA) as a prognostic predictor for stage I-III non-small cell lung cancer (NSCLC) patients with baseline tissue EGFR mutations. Transl Lung Cancer Res 10:3213–3225. https://doi.org/10.21037/tlcr-21-530
Ding X, Li L, Tang C, Meng C, Xu W, Wei X, Guo Z, Zhang T, Fu Y, Zhang L, Wang X, Lin L, Liang J (2018) Cytoplasmic expression of estrogen receptor β may predict poor outcome of EGFR-TKI therapy in metastatic lung adenocarcinoma. Oncol Lett 16:2382–2390. https://doi.org/10.3892/ol.2018.8936
Hitij NT, Kern I, Sadikov A, Knez L, Stanič K, Zwitter M, Cufer T (2017) Immunohistochemistry for EGFR mutation detection in non-small-cell lung cancer. Clin Lung Cancer 18:e187–e196. https://doi.org/10.1016/j.cllc.2016.11.021
Rudenko NV, Karatovskaya AP, Noskov AN, Shepelyakovskaya AO, Shchannikova MP, Loskutova IV, Artyemieva OA, Nikanova DA, Gladyr EA, Brovko FA (2018) Immunochemical assay with monoclonal antibodies for detection of staphylococcal enterotoxin H. J Food Drug Anal 26:741–750. https://doi.org/10.1016/j.jfda.2017.10.011
Zaini J, Syahruddin E, Yunus M, Andarini SL, Hudoyo A, Masykura N, Yasril R, Ridwanuloh A, Hidajat H, Nurwidya F, Suharsono S, Utomo ARH (2019) Evaluation of PCR-HRM, RFLP, and direct sequencing as simple and cost-effective methods to detect common EGFR mutations in plasma cell-free DNA of non-small cell lung cancer patients. Cancer Rep 2:e1159. https://doi.org/10.1002/cnr2.1159
Wang CC, Chen CA, Jong YJ, Kou HS (2018) Specific gene capture combined with restriction-fragment release for directly fluorescent genotyping of single-nucleotide polymorphisms in diagnosing spinal muscular atrophy. Anal Chem 90:11599–11606. https://doi.org/10.1021/acs.analchem.8b02996
Zhu D, Xing D, Shen X, Liu J, Chen Q (2004) High sensitive approach for point mutation detection based on electrochemiluminescence. Biosens Bioelectron 20:448–453. https://doi.org/10.1016/j.bios.2004.02.029
Zhao W, Liu M, Li H, Wang S, Tang S, Kong RM, Yu R (2019) Ultra-sensitive label-free electrochemical detection of the acute leukaemia gene Pax-5a based on enzyme-assisted cycle amplification. Biosens Bioelectron 143:111593. https://doi.org/10.1016/j.bios.2019.111593
Jiang D, Malla S, Fu YJ, Choudhary D, Rusling JF (2017) Direct LC-MS/MS detection of guanine oxidations in Exon 7 of the p53 tumor suppressor gene. Anal Chem 89:12872–12879. https://doi.org/10.1021/acs.analchem.7b03487
Chen YL, Jong YJ, Ferrance J, Hsien JS, Shih CJ, Feng CH, Wu MT, Wu SM (2008) Single nucleotide polymorphism detection in the hMSH2 gene using conformation-sensitive CE. Electrophoresis 29:634–640. https://doi.org/10.1002/elps.200700488
Kawada I, Soejima K, Watanabe H, Nakachi I, Yasuda H, Naoki K, Kawamura M, Eguchi K, Kobayashi K, Ishizaka A (2008) An alternative method for screening EGFR mutation using RFLP in non-small cell lung cancer patients. J Thorac Oncol 3:1096–1103. https://doi.org/10.1097/JTO.0b013e318186fadd
Cheng CW, Feng WH, Tang WH, Kou HS, Wang CC, Wu SM (2022) A three-step stacking capillary electrophoresis of field-amplified sample injection, sweeping, and micellar collapse for determination of dabigatran and its active metabolite in human plasma. J Food Drug Anal 30:88–103. https://doi.org/10.38212/2224-6614.3391
Liu TL, Fang LS, Liou JR, Dai JS, Chen YL (2021) Determination of quetiapine and its metabolites in plasma by field-enhanced sample stacking. J Food Drug Anal 29:709–716. https://doi.org/10.38212/2224-6614.3378
Wang M, Liu JK, Gao T, Xu LL, Zhang XX, Nie JH, Li Y, Chen HX (2022) A platform method for plasmid isoforms analysis by capillary gel electrophoresis. Electrophoresis 43:1174–1182. https://doi.org/10.1002/elps.202100343
Kang T, Lee H, Choe D, Joo SW, Lee SY, Yoon KA, Lee K (2012) Simultaneous detection of multiple mutations in epidermal growth factor receptor based on fluorescence quenching of quantum dots. Biosens Bioelectron 31:558–561. https://doi.org/10.1016/j.bios.2011.10.025
Tang Y, Zhang S, Wen Q, Huang H, Yang P (2015) A sensitive electrochemiluminescence cytosensor for quantitative evaluation of epidermal growth factor receptor expressed on cell surfaces. Anal Chim Acta 881:148–154. https://doi.org/10.1016/j.aca.2015.04.008
Tabata M, Liu X, Khamhanglit C, Kotaki S, Miyahara Y (2022) Detection of epidermal growth factor receptor expression in breast cancer cell lines using an ion-sensitive field-effect transistor in combination with enzymatic chemical signal amplification. J Am Chem Soc 144:16545–16552. https://doi.org/10.1021/jacs.2c06122
Zhou C, Li W, Zhao Y, Gu K, Liao Z, Guo B, Huang Z, Yang M, Wei H, Ma P, Li C, Li H, Tang Y, Lei C, Wang H (2023) Sensitive detection of viable salmonella bacteria based on tertiary cascade signal amplification via splintR ligase ligation-PCR amplification-CRISPR/Cas12a cleavage. Anal Chim Acta 1248:340885. https://doi.org/10.1016/j.aca.2023.340885
Zhou S, Sun H, Dong J, Lu P, Deng L, Liu Y, Yang M, Huo D, Hou C (2023) Highly sensitive and facile microRNA detection based on target triggered exponential rolling-circle amplification coupling with CRISPR/Cas12a. Anal Chim Acta 1265:341278. https://doi.org/10.1016/j.aca.2023.341278
Li Y, Wang J, Wang S, Wang J (2020) Rolling circle amplification based colorimetric determination of Staphylococcus aureus. Mikrochim Acta 187:119. https://doi.org/10.1007/s00604-019-4082-5
Li M, Xu X, Cai Q, Luo X, Zhou Z, Xu G, Xie Y (2019) Graphene oxide-based fluorometric determination of microRNA-141 using rolling circle amplification and exonuclease III-aided recycling amplification. Mikrochim Acta 186:531. https://doi.org/10.1007/s00604-019-3676-2
Wang CC, Chao KH, Chen YL, Chang JG, Wu SM (2012) Capillary electrophoretic genotyping of epidermal growth factor receptor for pharmacogenomic assay of lung cancer therapy. J Chromatogr A 1256:276–279. https://doi.org/10.1016/j.chroma.2012.07.087
Severins I, Szczepaniak M, Joo C (2018) Multiplex single-molecule DNA barcoding using an oligonucleotide ligation assay. Biophys J 115:957–967. https://doi.org/10.1016/j.bpj.2018.08.013
Sun Y, Lu X, Su F, Wang L, Liu C, Duan X, Li Z (2015) Real-time fluorescence ligase chain reaction for sensitive detection of single nucleotide polymorphism based on fluorescence resonance energy transfer. Biosens Bioelectron 74:705–710. https://doi.org/10.1016/j.bios.2015.07.028
Roth TL, Milenkovic L, Scott MP (2014) A rapid and simple method for DNA engineering using cycled ligation assembly. PLoS One 9:e107329. https://doi.org/10.1371/journal.pone.0107329
Zhang Y, Guo Y, Quirke P, Zhou D (2013) Ultrasensitive single-nucleotide polymorphism detection using target-recycled ligation, strand displacement and enzymatic amplification. Nanoscale 5:5027–5035. https://doi.org/10.1039/c3nr01010d
Li SH, Wu MH, Wang HM, Hsu PC, Fang YF, Wang CL, Chu HC, Lin HC, Lee LY, Wu CY, Yang CT, Chen JS, Hsieh JC (2022) Circulating EGFR mutations in patients with lung adenocarcinoma by circulating tumor cell isolation systems: a concordance study. Int J Mol Sci 23:10661. https://doi.org/10.3390/ijms231810661
Han S, Guo Y, Luo X, Tong G, Zhao C, Li Y, Guo T, Wang L, Gao N, Liu Y, Li H, Yang W (2022) Clinical value of alveolar lavage supernatant specimens in the detection of the EGFR gene mutation in patients with non-small cell lung carcinoma. Transl Cancer Res 11:1188–1194. https://doi.org/10.21037/tcr-22-681
Acknowledgements
We deeply extend our sincere thanks to volunteers and families who kindly contributed samples that were crucial to this study.
Funding
We gratefully acknowledge the Ministry of Science and Technology of Taiwan (MOST 108-2113-M-037-018-MY2) and the Zuoying Branch of Kaohsiung Armed Forces General Hospital (ZBH108-10) who provided financial assistance for our research and the Kaohsiung Medical University Chung-Ho Memorial Hospital, Kaohsiung, Taiwan, who offered the opportunities for collaboration in clinical application.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kou, HS., Lin, KH., Sebuyoya, R. et al. Dual-probe ligation without PCR for fluorescent sandwich assay of EGFR nucleotide variants in magnetic gene capture platform. Microchim Acta 190, 375 (2023). https://doi.org/10.1007/s00604-023-05950-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-023-05950-5