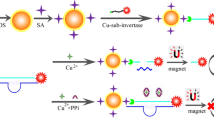
Abstract
A sensitive and portable biosensor is proposed for simple detection of microRNAs based on a supersandwich hybridization signal amplification strategy and a glucometer transducer. The presence of a target microRNA triggers the cascading hybridization chain reaction to create long supersandwich assemblies containing multiple biotin-labelled DNA probes. Then, large amounts of biotin-modified invertase signal molecules can attach to the supersandwich assemblies to generate an amplified signal for the glucometer readout. With such supersandwich format, a single target microRNA can introduce many biotin-invertase signal molecules, resulting in a one-to-multiple amplification effect. Thus, the accurate quantification of microRNAs can be achieved in a simple detection fashion without the requirement of expensive or precise instrumentation. The linear range of the biosensor for microRNA was from 0.05 to 100 nM with a detection limit of 48 pM. The proposed biosensor can discriminate the target microRNA from its family members with high selectivity and can be successfully applied to the detection of target microRNA spiked in serum samples with a good recovery (96.0–108.0%). Therefore, the proposed biosensor is expected to provide more information for early and accurate cancer diagnosis.
Graphical Abstract







Similar content being viewed by others
References
Jiang Z, Jiao J, Li J, Zhang H, Zheng J (2021) Novel electrochemical biosensing platform for microRNA: bivalent recognition-induced nanoparticle amplification occurred in nanochannels. Sens Actuators B 344:130209. https://doi.org/10.1016/j.snb.2021.130209
Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6:857–866. https://doi.org/10.1038/nrc1997
He X, Zeng T, Li Z, Wang G, Ma N (2016) Catalytic molecular imaging of microRNA in living cells by DNA-programmed nanoparticle disassembly. Angew Chem 55:3073–3076. https://doi.org/10.1002/anie.201509726
Li B, Liu Y, Liu Y, Tian T, Yang B, Huang X, Liu J, Liu B (2020) Construction of dual-color probes with target-triggered signal amplification for in situ single-molecule imaging of microRNA. ACS Nano 14:8116–8125. https://doi.org/10.1021/acsnano.0c01061
Xu F, Qiao Z, Luo L, He X, Lei Y, Tang J, Shi H, Wang K (2022) A label-free cyclic amplification strategy for microRNA detection by coupling graphene oxide-controlled adsorption with superlong poly(thymine)-hosted fluorescent copper nanoparticles. Talanta 243:123323. https://doi.org/10.1016/j.talanta.2022.123323
Liang J, Xu Q, Gu S (2022) LncRNA RSU1P2-microRNA let-7a-testis-expressed protein 10 axis modulates tumorigenesis and cancer stem cell-like properties in liver cancer. Bioengineered 13(2):4285–4300. https://doi.org/10.1080/21655979.2022.2031394
Shi K, Dou B, Yang C, Chai Y, Yuan R, Xiang Y (2015) DNA-fueled molecular machine enables enzyme-free target recycling amplification for electronic detection of microRNA from cancer cells with highly minimized background noise. Anal Chem 87(16):8578–8583. https://doi.org/10.1021/acs.analchem.5b02418
Li M, Cheng J, Yuan Z, Shen Q, Fan Q (2021) DNAzyme-catalyzed etching process of Au/Ag nanocages visualized via dark-field imaging with time elapse for ultrasensitive detection of microRNA. Sens. Actuators B 330:129347. https://doi.org/10.1016/j.snb.2020.129347
Peng H, Newbigging AM, Reid MS, Uppal JS, Xu J, Zhang H, Le XC (2020) Signal amplification in living cells: a review of microRNA detection and imaging. Anal Chem 92:292–308. https://doi.org/10.1021/acs.analchem.9b04752
Tang Y, He X, Zhou Z, Tang J, Guo R, Feng X (2016) Highly sensitive and selective miRNA detection based on a closed ring probe and multiple signal amplification. Chem Commun 52:13905–13908. https://doi.org/10.1039/c6cc07719f
Wu H, Wang H, Liu Y, Wu J, Zou P (2019) Fluorometric determination of microRNA by using target-triggered cascade signal amplification and DNA-templated silver nanoclusters. Mikrochim Acta 186:669. https://doi.org/10.1007/s00604-019-3789-7
Luo X, Zhu J, Jia W, Fang N, Wu P, Cai C, Zhu JJ (2021) Boosting long-range surface-enhanced raman scattering on plasmonic nanohole arrays for ultrasensitive detection of MiRNA. ACS Appl Mater Interfaces 13:18301–18313. https://doi.org/10.1021/acsami.1c01834
Li H, Li Y, Li W, Cui L, Huang G, Huang J (2020) A carbon nanoparticle and DNase I-assisted amplified fluorescent biosensor for miRNA analysis. Talanta 213:120816. https://doi.org/10.1016/j.talanta.2020.120816
Zou L, Wu Z, Liu X, Zheng Y, Mei W, Wang Q, Yang X, Wang K (2020) DNA hydrogelation-enhanced imaging ellipsometry for sensing exosomal microRNAs with a tunable detection range. Anal Chem 92:11953–11959. https://doi.org/10.1021/acs.analchem.0c02345
Kroh EM, Parkin RK, Mitchell PS, Tewari M (2010) Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 50:298–301. https://doi.org/10.1016/j.ymeth.2010.01.032
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294:853–858. https://doi.org/10.1126/science.1064921
Roy S, Soh JH, Ying JY (2016) A microarray platform for detecting disease-specific circulating miRNA in human serum. Biosens Bioelectron 75:238–246. https://doi.org/10.1016/j.bios.2015.08.039
Xu M, Ye J, Yang D, Abdullah Al-Maskri AA, Hu H, Jung C, Cai S, Zeng S (2019) Ultrasensitive detection of miRNA via one-step rolling circle-quantitative PCR (RC-qPCR). Anal Chim Acta 1077:208–215. https://doi.org/10.1016/j.aca.2019.05.028
Xian Y, Lu Y (2011) Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat Chem 3:697–703. https://doi.org/10.1038/nchem.1092
Xiang Y, Lu Y (2012) Using commercially available personal glucose meters for portable quantification of DNA. Anal Chem 84(4):1975–1980. https://doi.org/10.1021/ac203014s
Xiang Y, Lu Y (2012) Portable and quantitative detection of protein biomarkers and small molecular toxins using antibodies and ubiquitous personal glucose meters. Anal Chem 84(9):4174–4178. https://doi.org/10.1021/ac300517n
Li R, Liu Q, Jin Y, Li B (2020) Sensitive colorimetric determination of microRNA let-7a through rolling circle amplification and a peroxidase-mimicking system composed of trimeric G-triplex and hemin DNAzyme. Mikrochim Acta 187:139. https://doi.org/10.1007/s00604-019-4093-2
Qu X, Jin H, Liu Y, Sun Q (2018) Strand displacement amplification reaction on quantum dot-encoded silica bead for visual detection of multiplex microRNAs. Anal Chem 90:3482–3489. https://doi.org/10.1021/acs.analchem.7b05235
Zhou F, Li B, Ma J (2015) A linear DNA probe as an alternative to a molecular beacon for improving the sensitivity of a homogenous fluorescence biosensing platform for DNA detection using target-primed rolling circle amplification. RSC Adv 5:4019–4025. https://doi.org/10.1039/c4ra14467h
Zhou F, Meng R, Liu Q, Jin Y, Li B (2016) Photoinduced electron transfer-based fluorescence quenching combined with rolling circle amplification for sensitive detection of microRNA. ChemistrySelect 1:6422–6428. https://doi.org/10.1002/slct.201601485
Borghei Y-S, Hosseini M, Ganjali MR (2017) Fluorometric determination of microRNA via FRET between silver nanoclusters and CdTe quantum dots. Mikrochim Acta 184:4713–4721. https://doi.org/10.1007/s00604-017-2512-9
Xia F, White RJ, Zuo X, Patterson A, Xiao Y, Kang D, Gong X, Plaxco KW, Heeger AJ (2010) An electrochemical supersandwich assay for sensitive and selective DNA detection in complex matrices. J Am Chem Soc 132:14346–14348. https://doi.org/10.1021/ja104998m
Feng Q, Wang M, Qin L, Wang P (2019) Dual-signal readout of DNA methylation status based on the assembly of a supersandwich electrochemical biosensor without enzymatic reaction. ACS Sens 4:2615–2622. https://doi.org/10.1021/acssensors.9b00720
Liu H, Xu S, He Z, Deng A, Zhu J (2013) Supersandwich cytosensor for selective and ultrasensitive detection of cancer cells using aptamer-DNA concatamer-quantum dots probes. Anal Chem 85(6):3385–3392. https://doi.org/10.1021/ac303789x
Li J, Jiang J, Su Y, Liang Y, Zhang C (2021) A novel cloth-based supersandwich electrochemical aptasensor for direct, sensitive detection of pathogens. Anal Chim Acta 1188:339176. https://doi.org/10.1016/j.aca.2021.339176
Yang J, Huang X, Gan C, Yuan R, Xiang Y (2019) Highly specific and sensitive point-of-care detection of rare circulating tumor cells in whole blood via a dual recognition strategy. Biosens Bioelectron 143:111604. https://doi.org/10.1016/j.bios.2019.111604
Zhang J, Tang Y, Teng L, Lu M, Tang D (2015) Low-cost and highly efficient DNA biosensor for heavy metal ion using specific DNAzyme-modified microplate and portable glucometer-based detection mode. Biosens Bioelectron 68:232–238. https://doi.org/10.1016/j.bios.2015.01.001
Gao Y, Yu H, Tian J, Xiao B (2021) Nonenzymatic DNA-based fluorescence biosensor combining carbon dots and graphene oxide with target-induced DNA strand displacement for microRNA detection. Nanomaterials (Basel) 11(10):2608. https://doi.org/10.3390/nano11102608
Ma Q, Li S (2021) Enzyme- and label-free fluorescence microRNA biosensor based on the distance-dependent photoinduced electron transfer of DNA/Cu nanoparticles. Microchem J 160:105646. https://doi.org/10.1016/j.microc.2020.105646
Chen X, Wang J, Shen H, Su X, Cao Y, Li T, Gan N (2019) Microfluidic chip for multiplex detection of trace chemical contaminants based on magnetic encoded aptamer probes and multibranched DNA nanostructures as signal tags. ACS Sens 4(8):2131–2139. https://doi.org/10.1021/acssensors.9b00963
Funding
This work was supported by the National Natural Science Foundation of China (No. 21961046, No. 21362046, and No. 21062030), Yunnan Fundamental Research Projects (grant NO. 202201AU070056), Scientific Research Foundation Project of Yunnan Provincial Department of Education (No. 2021J0436), and PhD Scientific Research Foundation of Yunnan Normal University (No. 2020ZB009), College students’ Innovative Entrepreneurial Training plan program of Yunnan Province (No. S202110681054), Graduate Students’ Scientific Research Innovation Foundation of Yunnan Normal University (No. YJSJJ22-B64).
Author information
Authors and Affiliations
Contributions
LW: conceptualization, investigation, formal analysis, writing—original draft preparation.
TS: investigation, formal analysis.
LP: investigation, formal analysis.
JZ: software, validation.
RH: investigation, formal analysis, visualization.
YY: conceptualization, supervision.
JY: writing—reviewing and editing, supervision, funding acquisition.
YZ: writing—reviewing and editing, supervision, funding acquisition.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, L., Shan, T., Pu, L. et al. Glucometer-based electrochemical biosensor for determination of microRNA (let-7a) using magnetic-assisted extraction and supersandwich signal amplification. Microchim Acta 189, 444 (2022). https://doi.org/10.1007/s00604-022-05544-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-022-05544-7