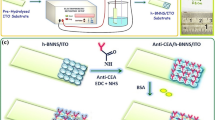
Abstract
Rapid and selective sensing of KRAS gene mutation which plays a crucial role in the development of colorectal, pancreatic, and lung cancers is of great significance in the early diagnosis of cancers. In the current study, we developed a simple electrochemical biosensor by differential pulse voltammetry technique for the specific detection of KRAS mutation that uses the mismatch-specific cleavage activity of T7-Endonuclease I (T7EI) coupled with horseradish peroxidase (HRP) to catalyze the oxidation of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate in the presence of hydrogen peroxide (H2O2). In addition, we synthesized the nanocomposite composed of multi-walled carbon nanotube/chitosan-ionic liquid/gold nanoparticles (MWCNT/Chit-IL/AuNPs) on screen-printed carbon electrode surface to increase the electrode surface area and electrochemical signal. In principle, T7E1 enzyme recognized and cleaved the mismatched site formed by the presence of KRAS gene mutation, removing 5′-biotin of capture probes and subsequently reducing the differential pulse voltammetry signal compared to wild-type KRAS gene. With this proposed strategy, a limit of detection of 11.89 fM was achieved with a broad linear relationship from 100 fM to 1 µM and discriminated 0.1% of mutant genes from the wild-type target genes. This confirms that the developed biosensor is a potential platform for the detection of mutations in early disease diagnosis.
Graphical abstract






Similar content being viewed by others
References
Liu P, Wang Y, Li X (2019) Targeting the untargetable KRAS in cancer therapy. Acta Pharm Sin B 9(5):871–879. https://doi.org/10.1016/j.apsb.2019.03.002
Porru M, Pompili L, Caruso C, Biroccio A, Leonetti C (2018) Targeting KRAS in metastatic colorectal cancer: current strategies and emerging opportunities. J Exp Clin Cancer Res 37(1):57. https://doi.org/10.1186/s13046-018-0719-1
Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D (2009) Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer 9(7):489–499. https://doi.org/10.1038/nrc2645
Chiu HM, Chang LC, Hsu WF, Chou CK, Wu MS (2015) Non-invasive screening for colorectal cancer in Asia. Best Pract Res Clin Gastroenterol 29(6):953–965. https://doi.org/10.1016/j.bpg.2015.09.015
Chung EJ, Yeom SS, Bonhan K, Tae YL, Jeong HL, Yong S, Lim SB (2017) Rapid and accurate detection of KRAS mutations in colorectal cancers using the isothermal-based optical sensor for companion diagnostics. Oncotarget 8(48):83860–83871
Li Y, Monzo M, Moreno I, Martinez-Rodenas F, Hernandez R, Castellano JJ, Canals J, Han B, Munoz C, Navarro A (2020) KRAS mutations by digital PCR in circulating tumor cells isolated from the mesenteric vein are associated with residual disease and overall survival in resected colorectal cancer patients. Int J Colorectal Dis 35(5):805–813. https://doi.org/10.1007/s00384-020-03538-6
Cha BC, Park KS, Park JS (2020) Signature mRNA markers in extracellular vesicles for the accurate diagnosis of colorectal cancer. J Biol Eng 14(1). https://doi.org/10.1186/s13036-020-0225-9
Hashkavayi AB, Cha BS, Hwang SH, Kim J, Park KS (2021) Highly sensitive electrochemical detection of circulating EpCAM-positive tumor cells using a dual signal amplification strategy. Sensors Actuators B Chem 343130087. https://doi.org/10.1016/j.snb.2021.130087
Malhotra P, Anwar M, Nanda N, Kochhar R, Wig JD, Vaiphei K, Mahmood S (2013) Alterations in K-ras, APC and p53-multiple genetic pathway in colorectal cancer among Indians. Tumour Biol 34(3):1901–1911. https://doi.org/10.1007/s13277-013-0734-y
Lang AH, Drexel H, Geller-Rhomberg S, Stark N, Winder T, Geiger K, Muendlein A (2011) Optimized allele-specific real-time PCR assays for the detection of common mutations in KRAS and BRAF. J Mol Diagn 13(1):23–28. https://doi.org/10.1016/j.jmoldx.2010.11.007
Tommy N, Mostofa R, Lena F (2000) Ulf dF, Ralf M, Pal N (2000) Direct analysis of single-nucleotide polymorphism on doublestranded DNA by pyrosequencing. Biotechnol Appl Biochem 31:107–112. https://doi.org/10.1042/BA19990104
Kam Y, Rubinstein A, Nissan A, Halle D, Yavin E (2012) Detection of endogenous K-ras mRNA in living cells at a single base resolution by a PNA molecular beacon. Mol Pharm 9(3):685–693. https://doi.org/10.1021/mp200505k
Okumura A, Sato Y, Kyo M, Kawaguchi H (2005) Point mutation detection with the sandwich method employing hydrogel nanospheres by the surface plasmon resonance imaging technique. Anal Biochem 339(2):328–337. https://doi.org/10.1016/j.ab.2005.01.017
Zhang Y, Wang L, Luo F, Qiu B, Guo L, Weng Z, Lin Z, Chen G (2017) An electrochemiluminescence biosensor for Kras mutations based on locked nucleic acid functionalized DNA walkers and hyperbranched rolling circle amplification. Chem Commun (Camb) 53(20):2910–2913. https://doi.org/10.1039/c7cc00009j
Xiao Q, Feng J, Li J, Liu Y, Wang D, Huang S (2019) A ratiometric electrochemical biosensor for ultrasensitive and highly selective detection of the K-ras gene via exonuclease III-assisted target recycling and rolling circle amplification strategies. Anal Methods 11(32):4146–4156. https://doi.org/10.1039/c9ay01007f
He Y, Cheng L, Yang Y, Chen P, Qiu B, Guo L, Wang Y, Lin Z, Hong G (2020) Label-free homogeneous electrochemical biosensor for HPV DNA based on entropy-driven target recycling and hyperbranched rolling circle amplification. Sens Actuators B: Chem 320. https://doi.org/10.1016/j.snb.2020.128407
Yang X, Lv J, Yang Z, Yuan R, Chai Y (2017) A sensitive electrochemical aptasensor for thrombin detection based on electroactive co-based metal-organic frameworks with target-triggering NESA strategy. Anal Chem 89(21):11636–11640. https://doi.org/10.1021/acs.analchem.7b03056
Chang Y, Zhuo Y, Chai Y, Yuan R (2017) Host-guest recognition-assisted electrochemical release: its reusable sensing application based on DNA cross configuration-fueled target cycling and strand displacement reaction amplification. Anal Chem 89(16):8266–8272. https://doi.org/10.1021/acs.analchem.7b01272
Zhao C, Gao F, Weng S, Liu Q, Lin L, Lin X (2016) An electrochemical sensor based on DNA polymerase and HRP-SiO2 nanoparticles for the ultrasensitive detection of K-ras gene point mutation. RSC Adv 6(11):8669–8676. https://doi.org/10.1039/c5ra24737c
Fang X, Bai L, Han X, Wang J, Shi A, Zhang Y (2014) Ultra-sensitive biosensor for K-ras gene detection using enzyme capped gold nanoparticles conjugates for signal amplification. Anal Biochem 460:47–53. https://doi.org/10.1016/j.ab.2014.05.019
Sun N, Guo Q, Li X, Chen J, Liu X, Wong K-Y, Fang Z (2018) An isothermal single base extension based lateral flow biosensor and electrochemical assay for gene point mutation detection. Anal Methods 10(24):2863–2868. https://doi.org/10.1039/c8ay00746b
Gosselin D, Gougis M, Baque M, Navarro FP, Belgacem MN, Chaussy D, Bourdat AG, Mailley P, Berthier J (2017) Screen-printed polyaniline-based electrodes for the real-time monitoring of loop-mediated isothermal amplification reactions. Anal Chem 89(19):10124–10128. https://doi.org/10.1021/acs.analchem.7b02394
Hashkavayi AB, Raoof JB, Ojani R (2018) Preparation of epirubicin aptasensor using curcumin as hybridization indicator: competitive binding assay between complementary strand of aptamer and epirubicin. Electroanalysis 30(2):378–385. https://doi.org/10.1002/elan.201700645
Zhang X, Huang C, Jiang Y, Shen J, Geng P, Zhang W, Huang Q (2016) An electrochemical glycan biosensor based on a thionine-bridged multiwalled carbon nanotube/gold nanoparticle composite-modified electrode. RSC Adv 6(114):112981–112987. https://doi.org/10.1039/c6ra23710j
Leszek AD, Mirka P, Andrzej K, Barbara L (2009) Synthesis and characterization of carbon nanotubes decorated with gold nanoparticles. Acta Physica Polonica Series a. https://doi.org/10.12693/APhysPolA.118.483
Bai L, Yuan R, Chai Y, Yuan Y, Wang Y, Xie S (2012) Direct electrochemistry and electrocatalysis of a glucose oxidase-functionalized bioconjugate as a trace label for ultrasensitive detection of thrombin. Chem Commun (Camb) 48(89):10972–10974. https://doi.org/10.1039/c2cc35295h
Qin X, Guo W, Yu H, Zhao J, Pei M (2015) A novel electrochemical aptasensor based on MWCNTs–BMIMPF6 and amino functionalized graphene nanocomposite films for determination of kanamycin. Anal Methods 7(13):5419–5427. https://doi.org/10.1039/c5ay00713e
Murugesan SN, Connahs H, Matsuoka Y (2021) Gupta Md, Huq M, Gowri V, Monroe S, Deem KD, Werner T, Tomoyasu Y. Monteiro A. https://doi.org/10.1101/2021.03.01.429915
Sentmanat MF, Peters ST, Florian CP, Connelly JP, Pruett-Miller SM (2018) A survey of validation strategies for CRISPR-Cas9 editing. Sci Rep 8(1):888. https://doi.org/10.1038/s41598-018-19441-8
Chudi G, Sanjay K, Rebecca K, Amy E (2004) Changing the enzymatic activity of T7 endonuclease by mutations at the â-Bridge Site: alteration of substrate specificity profile and metal ion requirements by mutation distant from the catalytic domain. Biochemistry 43(14). https://doi.org/10.1021/bi036033j.
Nahm FS (2017) What the P values really tell us. Korean J Pain 30(4):241–242. https://doi.org/10.3344/kjp.2017.30.4.241
Griffiths P, Needleman J (2019) Statistical significance testing and p-values: defending the indefensible? A discussion paper and position statement. Int J Nurs Stud 99:103384. https://doi.org/10.1016/j.ijnurstu.2019.07.001
Abnous K, Danesh NM, Alibolandi M, Ramezani M, Taghdisi SM (2017) Amperometric aptasensor for ochratoxin A based on the use of a gold electrode modified with aptamer, complementary DNA, SWCNTs and the redox marker Methylene Blue. Microchim Acta 184(4):1151–1159. https://doi.org/10.1007/s00604-017-2113-7
Han K, Liu T, Wang Y, Miao P (2016) Electrochemical aptasensors for detection of small molecules, macromolecules, and cells. Rev Anal Chem 35(4):201–211. https://doi.org/10.1515/revac-2016-0009
Funding
This work was supported by a grant from the National Research Foundation of Korea (NRF), funded by the Korea government (Ministry of Science and ICT) (No. NRF-2020R1C1C1012275), Korea Institute of Energy Technology Evaluation and Planning (KETEP) and the Ministry of Trade, Industry and Energy (MOTIE, 20194010201900), and by Konkuk University Researcher Fund in 2021.
Author information
Authors and Affiliations
Contributions
Pinky Chowdhury: conceptualization, methodology, validation, formal analysis, investigation, writing—original draft. Byung Seok Cha: methodology, formal analysis, review. Seokjoon Kim: methodology, formal analysis. Eun Sung Lee: methodology, formal analysis. Taehwi Yoon: formal analysis. Jisu Woo: formal analysis. Ki Soo Park: writing—review and editing, funding acquisition, resources, supervision.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chowdhury, P., Cha, B.S., Kim, S. et al. T7 Endonuclease I-mediated voltammetric detection of KRAS mutation coupled with horseradish peroxidase for signal amplification. Microchim Acta 189, 75 (2022). https://doi.org/10.1007/s00604-021-05089-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-021-05089-1