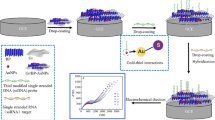
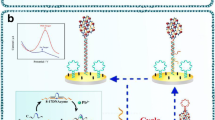
Abstract
An aptasensor for electrochemical detection of carbendazim is reported with mulberry fruit-like gold nanocrystal (MF-Au)/multiple graphene aerogel (MGA) and DNA cycle amplification. HAuCl4 was reduced by ascorbic acid in a CTAC solution containing KBr and KI and formed trioctahedron gold nanocrystal. The gold nanocrystal underwent structural evolution under enantioselective direction of l-cysteine. The resulting MF-Au shows a mulberry fruit-like nanostructure composed of gold nanocrystals of about 200 nm as the core and many irregular gold nanoparticles of about 30 nm as the shell. The exposure of high-index facets improves the catalytic activity of MF-Au. MF-Au/MGA was used for the construction of an aptasensor for electrochemical detection of carbendazim. The aptamer hybridizes with assistant strand DNA to form duplex DNA. Carbendazim binds with the formed duplex DNA to release assistant strand DNA, triggering one three-cascade DNA cycle. The utilization of a DNA cycle allows one carbendazim molecule to bring many methylene blue–labeled DNA fragments to the electrode surface. This promotes significant signal amplification due to the redox reaction of methylene blue. The detection signal is further enhanced by the catalysis of MF-Au and MGA towards the redox of methylene blue. A differential pulse voltammetric signal, best measured at − 0.32 V vs. Ag/AgCl, increases linearly with the carbendazim concentration ranging from 1.0 × 10−16 to 1.0 × 10−11 M with a detection limit of 4.4 × 10−17 M. The method provides ultrahigh sensitivity and selectivity and was successfully applied to the electrochemical detection of carbendazim in cucumber.
Graphical abstract

This study reports on an ultrasensitive aptasensor for electrochemical detection of carbendazim in cucumber based on mulberry fruit–like gold nanocrystal–multiple graphene aerogel and DNA cycle double amplification.
Similar content being viewed by others
Introduction
Carbendazim has been widely applied in fresh food production for the prevention of numerous pathogens and pests. However, extensive and unreasonable use of carbendazim may result in serious environmental pollution [1]. Researches demonstrated that carbendazim in the environment causes long-time accumulation in plants that absorb it through their roots and leaves [2]. A high level of carbendazim in food may cause adverse effects on the human body, such as endocrine disorders and cancers [3]. The maximum residue limit of carbendazim in cucumber is 0.1 mg kg−1 stipulated by the European Union. The USA has forbidden using carbendazim in agricultural production [4]. The development of a sensitive and selective method for the detection of carbendazim in fresh food is of importance.
The main analytical methods for the detection of carbendazim include high-performance liquid chromatography (HPLC) [5], gas chromatography-mass spectrometry (GC-MS) [6], surface-enhanced Raman scattering (SERS) [7], colorimetric method [8], fluorescent method [9], and electrochemical sensor [10]. HPLC and GC-MS are classical analytical methods for the detection of pesticides in the laboratory, with high sensitivity and selectivity, but these often require one complex sample pretreatment [11]. SERS offers a high sensitivity, but its instability of detection signal results in poor accuracy and repeatability [12]. The colorimetric method can use the naked eye to recognize the detection results and is suitable for on-site rapid carbendazim detection. However, its sensitivity is much lower than that of SERS, the fluorescent method, and the electrochemical sensor. This greatly limits its applications in the detection of carbendazim at low levels [13]. The fluorescent method has good sensitivity and specificity, but it often needs to use commercially unavailable luminescent materials as the optical probe for achieving response to carbendazim. In addition, the fluorescent methods based on common luminescent materials also suffer from the background fluorescence interference from biomacromolecules in food [1]. The electrochemical sensor has received more attention due to its ease of operation, high sensitivity, high selectivity, low cost, and portability [14].
To improve the sensitivity, some nanomaterials have been used as the sensing materials for electrochemical detection of insecticides such as carbon nanohorns [15], porous gold [16], copper [17], phosphorus-doped helical carbon fiber [18], and graphene [19]. Graphene and gold nanocrystal are the most frequently used sensing materials for the construction of electrochemical sensors due to their special property and structure. For example, Zhang et al. reported on one beta-cyclodextrin-graphene for the electrochemical detection of paichongding [20]. There was a linear relationship between peak current and pesticide concentration in the range from 1 to 10 μM. The detection limit reaches to 0.11 μM for the electrochemical detection of paichongding. Fei et al. reported on a composite composed of gold nanoparticle, multiwalled carbon nanotube, and reduced graphene oxide nanoribbon [21]. The composite has been successfully applied in the electrochemical detection of acetamiprid. Compared with common graphene, the sensor based on graphene aerogel exhibits an improved sensitivity for electrochemical detection. This is because graphene aerogel has a well-defined three-dimensional structure that can realize a high electron/ion conductivity [22]. However, the graphene aerogel prepared by using the traditional method is a kind of ultralight material composed of sparse graphene networks. The structural characteristic will greatly limit the improvement of electrical and mechanical properties of graphene. To resolve the problem, our group reported one strategy for the synthesis of multiple graphene aerogel (MGA) [23]. Such a MGA can achieve higher electron/ion conductivity and mechanical property compared with the common graphene aerogel, which further improves the sensitivity and repeatability for electrochemical detection. Different from graphene, gold nanocrystal possesses a special catalytic activity. In addition, gold nanocrystal can be also used to immobilize biomolecules on the electrode surface via the Au-S bond [24]. Compared with the low-index facets, high-index facets of gold nanocrystals have a better catalytic activity. Thus, the shape-controlled synthesis of gold nanocrystals with more exposed high-index facets becomes a new research hotspot [25]. Based on the above considerations, we have reason to believe that the combination of MGA with the gold nanocrystal with more exposed high-index facets could present a better electrochemical performance for the detection of carbendazim. However, there is no relevant research report up to now.
In the study, we reported on one strategy for synthesis of gold nanocrystals via the shape induction of CTAC, KBr, and KI as well as the enantioselective direction of l-cysteine. The as-synthesized gold nanocrystal (MF-Au) shows a mulberry fruit-like nanostructure with more exposed high-index facets. The aptasensor based on the combination of MF-Au with MGA and DNA cycle double amplification provides the advantages of sensitivity, specificity, and repeatability for the detection of carbendazim.
Experimental
Reagent and materials
Chloroauric acid (HAuCl4), cetyltrimethylammonium chloride (CTAC), potassium bromide (KBr), potassium iodide (KI), l-ascorbic acid, and l-cysteine were purchased from Sigma-Aldrich Co. LLC (Shanghai, China). Carbendazim and other pesticides were purchased from Beijing YiHua TongBiao Technology Co., Ltd. MGA was made by using a reported method [23]. All oligonucleotides used were synthesized and purified by Sangon Biotech. Co., Ltd. (Shanghai, China), and their base sequences are shown in Table s1. All DNA stock solutions were prepared in the Tris-EDTA (TE) buffer of pH 7.4, composed of 10 mM Tris-HCl, 12.5 mM MgCl2, and 1 mM ethylenediaminetetraacetic acid (EDTA), and stored at − 20 °C. Before use, the aptamer (AP), assistant strand DNA (AS-DNA), methylene blue–modified hairpin DNA1 (MB-H1), and hairpin DNA2 (H2) were separately pretreated by heating at 95 °C for 5 min. The restriction endonuclease (Nt.AlwI) and the cutsmart buffer (pH 7.9), composed of 50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, and 100 μg mL−1 bovine serum albumin (BSA), were purchased from New England Biolabs, Inc. (USA). Other reagents used were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Ultrapure water (18.2 MΩ cm) was used throughout the experiment.
MF-Au synthesis
The HAuCl4 solution (0.7 mL, 10 mM) was mixed with the mixed solution of 8.85 mL, composed of 17 mM CTAC, 5.65 μM KBr, and 0.57 μM KI. The l-ascorbic acid solution (0.45 mL, 50 mM) was rapidly injected into the above solution. The solution was incubated at 20 °C for 30 min to obtain gold seed solution.
The as-prepared Au seed solution (1.03 mL), CTAC solution (16.46 mL, 100 mM), KBr solution (1.03 mL, 1 mM), KI solution (103 μL, 1 mM), and the l-cysteine solution (103 μL, 1 mM) were mixed. Then, ultrapure water was added until the total volume reached 100 mL. The resulting solution of 4.86 mL was transferred into a centrifugal tube of 20 mL, followed by the addition of HAuCl4 (200 μL, 10 mM), injection of 475 μL of l-ascorbic acid (50 mM), incubation at 20 °C for 6 min, centrifugation at 4000 rpm for 1 min, and washing three times with ultrapure water in sequence. To obtain a MF-Au stock solution, the collected MF-Au was re-dispersed in 0.1 mL of ultrapure water. The above procedure was also used for synthesis of one gold nanomaterial control sample (Au-C) unless there was no addition of l-cysteine.
Sensor preparation
The MF-Au stock solution was mixed with an equal volume of MGA dispersion (1.0 mg mL−1) and then ultrasonicated for 30 min. To obtain MF-Au-MGA/GCE, its 5 μL was dropped on the glass carbon electrode surface (GCE, 2 mm in diameter) and incubated at room temperature overnight. Next, the linker DNA (L-DNA) solution (100 μL, 10 μM) was mixed with an equal volume of tris(2-carboxyethyl) phosphine hydrochloride (TCEP) solution (20 mM) and then stirred for 1 h. Its 5 μL was dropped on the MF-Au-MGA/GCE surface and incubated at 37 °C for 1 h. The resulting L-DNA/MF-Au-MGA/GCE was washed with ultrapure water to remove free L-DNA and dried with N2 current. To block the unmodified active sites on the surface of MF-Au, 5 μL of the 6-mercapto-1-hexanol (MCH) in the TE buffer (1 mM) was dropped on the L-DNA/MF-Au-MGA/GCE surface and then incubated at 37 °C for 1 h. The resulting MCH/L-DNA/MF-Au-MGA/GCE was washed with phosphate-buffered solution (PBS) of pH 7.4, dried with N2 current, and stored at 4 °C before use.
Sample pretreatment
All cucumber samples were collected from markets in Wuxi City, washed, and dried in room temperature. The resulting cucumber samples were cut into small pieces and then crushed into a paste in a household juice machine to form homogeneous cucumber juice. The juice of 10 g was placed into a 50-mL Teflon centrifuge tube and shaken with a wrist action shaker for 1 min; then, 6 g MgSO4, 1 g NaCl, and 1.5 g C6H5O7Na3·2H2O were added and the tube shaken immediately and vigorously for 1 min by vortex. The tubes were centrifuged at 4000 rpm for 5 min; solid residue was discarded and mixed with 2 mL of glacial acetic acid. It was treated by shaking in a vortex and centrifuging for 2 min at 4000 rpm [11]. The collected supernatants were used for the detection of carbendazim.
Carbendazim detection
The pretreated AS-DNA solution (1 μM) was mixed with an equal volume of pretreated AP solution (1 μM) and subsequently incubated at 37 °C for 2 h to form duplex DNA. The carbendazim standard solution of 20 μL or 40 μL of carbendazim sample solution was transferred into a centrifuge tube of 2 mL and incubated for 2 h at 37 °C. The pretreated MB-H1 (10 μM, 10 μL), pretreated H2 (10 μM, 10 μL), and cutsmart buffer (10 μL) were added then incubated at 37 °C for 20 min. Followed by the addition of Nt.AlwI solution (2 U μL−1, 5 μL), incubation for 1 h at 37 °C and heating at 80 °C for 10 min to deactivate the Nt.AlwI in the system.
The deactivated solution of 5 μL was dropped on the surface of prepared MCH/L-DNA/MF-Au-MGA/GCE, incubated at 37 °C for 1 h and washed with PBS (pH 7.4, 10 mM). The incubated electrode was immersed in PBS buffer of pH 7.4 and then the differential pulse voltammetry (DPV) at − 0.32 V vs. Ag/AgCl.
Results and discussion
Synthesis and characteristics
Synthesis of MF-Au includes two cumulative steps, namely preparation of gold seed and MF-Au. Firstly, gold seed was made via the reduction of HAuCl4 with ascorbic acid in the presence of CTAC, KBr, and KI. In the synthesis, CTAC was used as a surfactant to make the soft template. The template offers an independent space for the reduction of HAuCl4 and subsequent gold crystal growth. A relatively large volume of the template leads to the formation of a relatively gold nanocrystal with a particle size of hundreds of nanometers. In addition, CTAC was also used as a shape inducer to make trioctahedron gold seeds. The previous investigation has revealed that CTA+ can be selectively adsorbed on the surface of gold nanocrystals [26]. Due to a higher surface energy, the low-index facet of gold nanocrystals has a higher adsorption ability towards CTA+ and Cl− compared with the high-index facet. As more adsorption results in a greater decrease in the growth rate of crystal facets, the adsorption can effectively inhibit the growth of low-index facets and leads to the formation of polyhedral gold nanocrystals [27]. Both KBr and KI were used to improve the morphology uniformity of gold seeds. Although Cl−, Br−, and I− have similar chemical properties, there exists an obvious difference in their adsorption ability on the different crystal facets and corresponding kinetic behavior. As a result, the gold seed induced by different halogen anions may offer a different nanostructure [28]. To make full use of the complementarity of Cl−, Br−, and I− in the shape induction, the mixture of Cl−, Br−, and I− was adopted for the shape-controlled synthesis of gold seeds. The resulting gold seeds were characterized by SEM analysis shown in Fig. s1. The gold seeds exhibits a trioctahedron-like nanostructure. Next, the as-prepared gold seeds were grown in the presence of l-cysteine to finally produce MF-Au.
The as-synthesized MF-Au was characterized by SEM, TEM, element mapping, and XRD techniques. Figure 1 presents SEM, TEM, high-resolution TEM (HRTEM), electron image, elemental mappings, and EDS layered image of the MF-Au. The MF-Au shows a mulberry fruit-like nanostructure, composed of the gold nanocrystal with a particle size of about 200 nm as the core and many irregular gold nanocrystals with a particle size of about 30 nm as the shell. The integration of a large gold core with a shell composed of irregular gold nanoparticles simultaneously achieves high electrical conductivity, a large specific surface, and more exposed high-index facets. The large specific surface increases the contact area between the catalyst and the redox probe. The exposed high-index facets improve the catalytic activity [25]. The plane spacings of (200) and (111) facets on the HRTEM image were measured by the ImageJ software. The plane spacings are 0.22 ± 0.03 for the (200) facet and 0.28 ± 0.02 nm for the (111) facet. The elemental mapping images of the Au, I, Br, Cl, S, and N elements in MF-Au indicate that all elements exhibit a uniform distribution. This demonstrates that these elements were involved in the formation and growth of gold nanocrystals. Further, the EDS analysis was used to determine the element composition on the surface of MF-Au (Fig. s2 and Table s2). It was found from Table s2 that the atomic percentages of the Au, I, Br, Cl, S and N elements are 91.65, 0.2, 1.57, 4.14, 2.24, and 0.2. Since the S element only comes from l-cysteine, a relatively high S context proves a strong affinity between the gold nanocrystal and l-cysteine during the gold crystal growth process.
The XRD pattern of as-synthesized MF-Au in Fig. 2 includes eight strong XRD diffraction peaks at 38.17°, 44.47°, 64.65°, 77.64°, 81.80°, 98.13°, 110.91°, and 115.33°. These peaks correspond to the (111), (200), (220), (311), (222), (400), (331), and (420) crystal facets of gold crystal (JCPDS 4-0784). Figure 2 also indicates that the high-index facets of MF-Au such as the (331) and (420) display high diffraction intensities, which are obviously higher than that of the gold seeds (Fig. s3). The result verifies that the introduction of l-cysteine can improve the exposure of high-index facets. The improvement helps to enhance the catalytic activity and leads to a more sensitive electrochemical response [29].
To understand the role of l-cysteine in the synthesis of MF-Au, one gold nanocrystal control sample (Au-C) was prepared by using the same procedure but with no addition of l-cysteine. Its structure was characterized by SEM and TEM techniques and the results were shown in Fig. s4. Compared with the gold seed, the Au-C offers a larger particle size. During the growth process of gold seeds, a large number of Au3+ ions were reduced into Au0 atoms. These Au0 atoms finally deposited on the surface of the gold seeds and formed larger gold nanocrystals. However, the Au-C displays a similar morphology with the corresponding gold seed but different from that of the MF-Au. For the synthesis of Au-C and gold seed, the same soft template (CTAC) and shape inducers (CTAC, KBr, and KI) were used for the formation of gold nanocrystals. This will lead to the formation of trioctahedron-like morphologies in the Au-C and the gold seed. Different from the Au-C and gold seed, there are many irregular edges and corners on the crystal face of MF-Au. As for the synthesis of Au-C and MF-Au, the only difference between them is that l-cysteine was also used as a shape inducer in the synthesis of MF-Au; the formation of irregular edges and corners in MF-Au cold be attributed to the use of l-cysteine. l-Cysteine contains a chiral carbon atom and a sulfhydryl group. The enantioselective interaction on the interfaces caused by l-cysteine achieves asymmetric structural evolution of gold nanocrystals. This improves the exposure of high-index facets [25].
Sensing principle
The scheme for the construction of the aptasensor for electrochemical detection of carbendazim is presented in Fig. 3. Firstly, MF-Au and MGA powder were ultrasonically dispersed in ultrapure water to form a stable dispersion. The part of dispersion was drop-coated on the GCE surface to produce a MF-Au-MGA composite, in which MF-Au nanoparticles were evenly distributed on the graphene sheets (Fig. s5). Then, L-DNA was connected to the surface of MF-Au-MGA/GCE via the Au-S bond. Finally, MCH was used to block the active site on the surface of MF-Au to avoid nonspecific adsorption in the detection of carbendazim.
The as-prepared aptasensor coupled with DNA cycle amplification was applied in the electrochemical detection of carbendazim. In the detection, AP was firstly hybridized with AS-DNA to produce duplex DNA (AP-AS-DNA). Then, carbendazim binds with the formed duplex DNA to release AS-DNA. This results in triggering a three-cascade DNA recycle. The released AS-DNA was combined with MB-H1 to produce MB-H1-AS-DNA duplex DNA that contains one restriction endonuclease Nt.AlwI recognition site. Nt.AlwI can cleavage H1 sequence at the recognition site in MB-H1-AS-DNA to produce a secondary trigger strand DNA (STS-DNA) and methylene blue–modified signal strand DNA (MB-SS-DNA), leading to the release of AS-DNA. The released AS-DNA interacts with MB-H1 to initiate the next DNA recycle and produces many MB-SS-DNA (cycle I). Meanwhile, the formed STS-DNA will specifically bind with H2 to produce H2-STS-DNA with the Nt.AlwI recognition site. Nt.AlwI cleavages H2-STS-DNA at the recognition site to release STS-DNA and AS-DNA. The released STS-DNA was directly used for the next cleavage cycle of H2, triggering cycle II. The released AS-DNA in cycle II was directly used to trigger cycle I and initiates one new DNA recycling (cycle III). By the three-cascade recycle, one carbendazim molecule can produce many MB-SS-DNA probes, which were combined with the L-DNA immobilized on the electrode surface by base complementary pairing. The redox of these MB probes on the electrode surface brings a significant signal amplification. The detection signal was further enhanced by the in situ catalysis of MF-Au towards the redox of MB due to the excellent catalytic activity of MF-Au.
To understand the sensing principle, cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS) behaviors of different electrodes were studied in 1 mM Fe(CN)64− in PBS of pH 7.4. As shown in Fig. 4, all CV curves contain one pair of redox peaks, which are attributed to the oxidization and reduction of Fe(CN)6]3−/4− on the electrode surface. The immobilization of MF-Au-MGA results in an increased CV current density with a reduced charge transfer impedance (Rct). The increase is due to high electron/ion conductivity of MF-Au-MGA. The high conductivity accelerates the electrode reaction and brings a more sensitive CV current response. The current density will rapidly increase with the increase of the scan rate, and the peak current has a linear relationship with the square root of the scan rate (Fig. s6). This verifies that the redox of [Fe(CN)6]3−/4 on the electrode surface is a quasi-reversible electrochemical process. However, the CV current will decrease after the L-DNA was connected on the MF-Au-MGA/GCE with the increase of the Rct value. This is because the L-DNA partly blocks the channels for electron transfer between the electrode and electrolyte. In addition, the negative charge of the DNA backbone is responsible for an impeded electron transfer, assuming electrostatic repulsion between the negatively charged L-DNA molecule and negatively charged [Fe(CN)6]3−/4− probes. This further reduces the CV current response. Similar to L-DNA, further MCH immobilization brings a decrease in the CV current density. To examine the response towards carbendazim, the as-prepared aptasensor was incubated in a mixed reaction solution composed of AP, AS-DNA, MB-H1, H2, and Nt.AlwI for 1 h in the presence of carbendazim. After that, its CV and EIS curves were measured. The result shows that the incubation further decreases the CV current density with increasing Rct value. This is because in the presence of carbendazim one carbendazim molecule brings many methylene blue-labeled DNA fragments to the electrode surface by the DNA cycle. The poor electrical conductivity of the DNA fragments on the electrode surface leads to an increase in Rct and a reduced CV current density. More importantly, the above result demonstrates that carbendazim can trigger the three-cascade recycle and achieves a sensitive electrochemical response. Therefore, the proposed aptasensor can be applied in the electrochemical detection of carbendazim.
CV curves (A) and EIS curves (B) in 1.0 mM Fe(CN)64− in 10 mM PBS (pH 7.4) of the bare GCE (a), MF-Au-MGA/GCE (b), L-DNA/MF-Au-MGA/GCE (c), and MCH/L-DNA/MF-Au-MGA/GCE before (d) and after incubation in a mixed reaction solution for 1 h in the presence of 1 × 10−12 M carbendazim (e). Insert: the equivalent circuit for EIS
Electrochemical response
To examine the electrochemical response towards carbendazim, the DPV curves of the aptasensor before and after incubation in the mixed reaction solution composed of AP, AS-DNA, MB-H1, H2, Nt.AlwI, and carbendazim were measured in PBS of pH 7.4. Figure 5 shows that in the absence of carbendazim, there is a very weak peak on the DPV curve. The weak DPV signal may be due to the background current produced by the measurement system. However, the incubation causes an obviously increased peak current in the presence of carbendazim. This is because the presence of carbendazim triggers the three-cascade DNA cycle. By the DNA cycle, many MB redox probes were immobilized on the electrode surface. The redox of these MB molecules produces a high DPV current response.
Four reaction systems were designed to evaluate the roles of important reagents in the DNA recycle, including the reaction system lacking only MB-H1 (system I), the reaction system lacking only Nt.AlwI (system II), the reaction system lacking only H2 (system III), and the complete reaction system (system IV). For each system, the aptasensor was incubated for 1 h in the presence of 1 × 10−12 M carbendazim and then used for the DPV measurement in PBS of pH 7.4. Figure 6 shows the aptasensors incubated in system I and system II give a small DPV peak current (Ip). This is because in the absence of MB-H1 or Nt.AlwI the three-cascade cycle cannot be triggered. As no MB probes were brought to the electrode surface, the aptasensor gives a low Ip value due to the lack of any redox probes on the electrode surface. However, the aptasensor incubated in system III offers an obviously increased Ip value. In the absence of H2, only cycle I can be achieved. By cycle I, one carbendazim molecule can bring many MB redox probes to the electrode surface. The redox of these MB probes produces a sensitive DPV current response. When system III was replaced by system IV, the Ip value will be further enhanced. For system IV, the three-cascade recycle can be triggered by carbendazim. As the three-cascade DNA recycle has a much higher efficiency to produce MB-SS-DNA sequences compared with single cycle I, the aptasensor incubated in system IV exhibits a much higher Ip value than that in system III due to more MB redox probes on the electrode surface.
In the detection, only a tiny fraction of L-DNA molecules on the electrode surface was bound by MB-SS-DNA. Thus, the proposed aptasensor can be directly used for the next detection of carbendazim after the detection was completed. To evaluate the reusability of the aptasensor, the aptasensor was repeatedly used to measure 1 × 10−12 M carbendazim by using the same procedure. The results showed that each of the detections gives a similar ΔIp value when the detection time is less than 120, indicating a good reusability.
Condition optimization
To optimize the conditions for electrochemical detection of carbendazim, the effects of various factors on the DPV current response towards 1 × 10−12 M carbendazim were investigated. Fig. s7 presents the relationship curves of the Ip value with the Nt.Alwl concentration and the cleavage time. If the cleavage time was fixed at 60 min, the Ip value rapidly increased with increasing Nt.AlwI concentration when the concentration was less than 1.2 U μL−1. However, the increasing trend of the Ip value slowed down when the Nt.AlwI concentration was more than 1.2 U μL−1, reached the maximum at 1.5 U μL−1, and then tended to be stable with the increase of Nt.Alwl concentration. If the Nt.AlwI concentration was fixed at 2 U μL−1, the Ip value increased with the increase of cleavage time when the cleavage time was less than 45 min. The value remained almost unchanged after 45 min. This is because the remaining H1 and H2 in the reaction system will rapidly decrease with the increase of cleavage time. The reduction will eventually lead to a decrease in the speed of MB probes being carried to the electrode surface. Consequently, the growth rate of the DPV peak current decreases with the increase of cleavage time. When the cleavage time is more than 45 min, the capture of MB probes by the electrode surface and the process of MB probes leaving the electrode surface reaches its equilibrium state. At the equilibrium state, the DPV current response could not be changed by prolonging the cleavage time because the number of MB molecules can remain almost unchanged. To obtain a high sensitivity and save the analysis time, the Nt.AlwI concentration of 2 U μL−1 and the cleavage time of 60 min were selected for electrochemical detection of carbendazim.
Relationship curves of the Ip values with the concentrations of AP, AS-DNA, MB-H1, and H2, and the incubation times for hybridization of AP with AS-DNA, dissociation of AS-DNA from the AP-AS-DNA duplex, and immobilization of MB-SS-DNA probes are successively presented in Fig. s8, Fig. s9, Fig. s10, Fig. s11, Fig. s12, Fig. s13, and Fig. s14. The optimized concentrations are 1 μM of AP, 1 μM of AS-DNA, 10 μM of MB-H1, and 10 μM of H2. The optimized times are 2 h for the hybridization of AP with AS-DNA, 2 h for the dissociation of AS-DNA from the AP-AS-DNA duplex, and 1 h for the immobilization of MB-SS-DNA probes on the electrode surface.
Analytical characteristics
Under the optimized conditions, the DPV current response of the proposed aptasensor towards different concentrations of carbendazim was investigated. Figure 7A indicates that the DPV current will increase with the increase of carbendazim. A higher concentration of carbendazim can combine with more AP probes in the AP-AS-DNA to release more AS-DNA molecules. This results in the production of more MB-SS-DNA probes by the three-cascade cycle. The formed MB-SS-DNA probes were immobilized on the electrode surface and produced a more sensitive DPV current response, indicating a bigger Ip value. Further, Fig. 7B indicates the calibration plot between the DPV peak current (Ip) and the logarithm of carbendazim concentration. The Ip value gives a good linear relationship with the logarithm of carbendazim concentration in the range of 1.0 × 10−16–1.0 × 10−11 M. The corresponding linear regression equation is Ip (nA) = 143.06 × LOG[CCBZ, M] + 2643.8, and the correlation coefficient is 0.992. The detection limit was 4.4 × 10−17 M (S/N = 3).
The DPV curves (A) in PBS of pH 7.4 of the proposed aptasensor incubated for 1 h in the presence of 0.0, 1.0 × 10−16, 5.0 × 10−15, 1.0 × 10−14, 5.0 × 10−13, 1.0 × 10−12, 5.0 × 10−11, 1.0 × 10−10, 5.0 × 10−9, and 1.0 × 10−8 M carbendazim (from bottom to top) and the calibration plot (B) between the Ip value at − 0.32 V vs. Ag/AgCl and the logarithm of carbendazim concentration
Twenty aptasensors were prepared by the same procedure and materials and then used for electrochemical detection of 1.0 × 10−12 M carbendazim. The DPV peak currents of twenty aptasensors are shown in Fig. s15. A relative standard deviation (RSD) of 1.7% was obtained from Fig. s15, indicating a good reproducibility. One aptasensor was repeatedly used for the detection of 1.0 × 10−12 M carbendazim for twenty times. The changes (ΔIp) in the DPV peak current caused by these detections are presented in Fig. s16. The RSD is about 2.3%, verifying a high repeatability. The aptasensor was stored at 4 °C for 30 days. It was taken out and used for the electrochemical detection of 1.0 × 10−12 M carbendazim every 2 days. The ΔIp values caused by the detection are shown in Fig. s17. The RSD for the storage period of 30 days is 2.5%. The result demonstrates that the proposed aptasensor has a long-term stability.
The inorganic ions and organic small-molecule compounds found in large quantities in biological samples and homologous pesticides commonly used in cucumber production in China were used to evaluate the anti-interference capability of the proposed aptasensor in the detection of carbendazim. Inorganic ions, including 1 × 10−9 M of Ca2+, Mg2+, Na+, K+, C2O42−, CO32−, SO42−, PO43−, and Cl−, and organic compounds, including 1 × 10−11 M of ascorbic acid, glucose, malic acid, citric acid, and similar compounds to carbendazim, including thiabendazole, 2-aminobenzimidazole, and fuberidazole, were used to replace carbendazim for the DPV measurement. Figure 8 shows that only carbendazim can bring a sensitive DPV current response. This is because the other substances used cannot trigger the three-cascade cycle process. As no MB probes were brought to the electrode surface after the incubation, the DPV peak currents caused by the other substances tested are very weak. Therefore, the as-proposed aptasensor has a good selectivity for electrochemical detection of carbendazim.
The DPV current (Ip) at − 0.32 V vs. Ag/AgCl in 10 mM PBS (pH 7.4) towards 1 × 10−9 M of Ca2+ (a), Mg2+ (b), Na+ (c), K+ (d), C2O42− (e), CO32− (f), SO42− (g), PO43− (h), Cl− (i), 1 × 10−11 M of ascorbic acid (j), glucose (k), malic acid (l), citric acid (m), thiabendazole (n) and 2-aminobenzimidazole (o), 1 × 10−12 M of carbendazim (p), and 1 × 10−11 M of fuberidazole (q)
For the sake of comparison, analytical parameters of the electrochemical sensors for the detection of carbendazim reported in literature are shown in Table 1. Table 1 shows that the proposed aptasensor gives the highest sensitivity among the reported methods. This is due to high catalytic activity of MF-Au unless DNA cycle amplification. Because of high catalytic activity, the catalytic effect of MF-Au on the redox of MB greatly improves the DPV response towards carbendazim. To evaluate the catalytic activity of MF-Au/MGA, DPV behaviors of the electrodes modified by different materials were investigated, including gold seed, MF-Au, MF-Au/graphene, MF-Au/MGA , and the spherical gold nanoparticles (Au-S) prepared by the same procedure used in the synthesis of gold seeds unless there was no addition of CTAC. The results are shown in Fig. 9. For the bare GCE, there is one peak on the DPV curve. This is due to the oxidation of the MB probe. The current will obviously increase when the Au-S was modified on the GCE surface. This demonstrates that the Au-S can offer the catalytic activity towards the MB probe. When the Au-S was replaced by the gold seed, the current increased. This confirmed that the gold seed had a better catalytic activity towards the MB probe compared with the Au-S. Different from the Au-S, the gold seed showed a trioctahedron-like nanostructure with more exposed high-index facets. The exposure improved the catalytic activity and accelerated the oxidation of the MB probe, leading to an increased current response. The CV current further increased when the gold seed was replaced by MF-Au. This demonstrated that the MF-Au can offer the highest catalytic activity among the three kinds of gold nanomaterials. This is because the MF-Au provides more exposed high-index facets compared with the gold seed. Interestingly, the combination of MF-Au with graphene brings a higher DPV current. In the MF-Au/graphene composite, a large graphene sheet acts as a particle connector to connect the isolated MF-Au particles into a whole. This further enhances the electrical conductivity and realizes a better catalytic activity. Compared with common graphene, the MGA creates a much higher electron/ion conductivity and mechanical property. As a result, the combination of MF-Au with MGA achieves the highest DPV current response. This helps to greatly improve the sensitivity and repeatability for electrochemical detection of carbendazim.
Sample analysis
The as-proposed aptasensor was applied in the electrochemical detection of carbendazim in cucumber samples. The recovery experiment was carried out by adding a known concentration of the standard carbendazim into the cucumber juice from the cucumber sample. The spiked concentration of carbendazim was based on the information with the experimental evidences [11]. Table 2 shows the recovery of carbendazim in the cucumber samples was in the range of 96–104%. The results of the proposed aptasensor are consistent with those obtained by the HPLC-MS method. These confirm that the proposed method has admirable accuracy and reliability for carbendazim analysis in real cucumber samples.
Conclusions
The study demonstrated one strategy for the synthesis of gold nanocrystal via the asymmetric structural evolution under the enantioselective direction of l-cysteine. The as-synthesized gold nanocrystal offers a mulberry fruit-like nanostructure. The unique structure has more exposed high-index facets and leads to an improved catalytic activity. The combination of mulberry fruit-like gold nanocrystal with multiple graphene aerogel was used as the sensing material for construction of the aptasensor for electrochemical detection of carbendazim coupled with the DNA cycle amplification. Significant double signal amplification provided by gold nanoparticles and DNA recycle achieves an ultrasensitive detection of carbendazim. The sensitivity is much better than that of all reported methods for the detection of carbendazim. The as-proposed aptasensor can also be used for the detection of other insecticides at a very low level in foods and environmental samples through the redesign of DNA sequences in DNA recycle.
References
Chen M, Zhao Z, Lan X, Chen Y, Zhang L, Ji R, Wang L (2015) Determination of carbendazim and metiram pesticides residues in reapeseed and peanut oils by fluorescence spectrophotometry. Measurement 73:313–317. https://doi.org/10.1016/j.measurement.2015.05.006
Feng SX, Li YG, Zhang RY, Li YC (2019) A novel electrochemical sensor based on molecularly imprinted polymer modified hollow N, S-Mo2C/C spheres for highly sensitive and selective carbendazim determination. Biosens Bioelectron 142:111491. https://doi.org/10.1016/j.bios.2019.111491
Petroni JM, Lucca BG, Fogliato DK, Ferreira VS (2016) Sensitive approach for voltammetric determination of carbendazim based on the use of an anionic surfactant. Electroanal 28:1362–1369. https://doi.org/10.1002/elan.201501069
Li Q, Dou XW, Zhao XS, Zhang L, Luo JY, Xing XY, Yang MH (2019) A gold/Fe3O4 nanocomposite for use in a surface plasmon resonance immunosensor for carbendazim. Microchim Acta 186:313. https://doi.org/10.1007/s00604-019-3402-0
Akkbik M, Hazer O (2018) Novel molecularly imprinted polymer for the determination of carbendazim from water and food by solid-phase extraction and high-performance liquid chromatography. Anal Lett 51:7–23. https://doi.org/10.1080/00032719.2017.1284859
Yang BC, Wan XD, Yang X, Li YJ, Zhang ZY, Wan XJ, Luo Y, Deng W, Wang F, Huang OP (2018) Rapid determination of carbendazim in complex matrices by electrospray ionization mass spectrometry with syringe filter needle. J Mass Spectrom 53:234–239. https://doi.org/10.1002/jms.4057
Furini LN, Samchez-Cortes S, Lopez-Tocon I, Otero JC, Aroca RF, Constantino CJL (2015) Detection and quantitative analysis of carbendazim herbicide on Ag nanoparticles via surface-enhanced Raman scattering. J Raman Spectrosc 46:1095–1101. https://doi.org/10.1002/jrs.4737
Ma Y, Jiang H, Shen C, Hou C, Huo D, Wu H, Yang M (2017) Detection of carbendazim residues with a colorimetric sensor based on gold nanoparticles. J Appl Spectrosc 84:460–465. https://doi.org/10.1007/s10812-017-0492-5
Yang Y, Xing XX, Zou T, Wang ZD, Zhao RJ, Hong P, Peng SJ, Zhang X, Wang YD (2020) A novel and sensitive ratiometric fluorescence assay for carbendazim based on N-doped carbon quantum dots and gold nanocluster nanohybrid. J Hazard Mater 386:121958. https://doi.org/10.1016/j.jhazmat.2019.121958
Eissa S, Zourob M (2017) Selection and characterization of DNA aptamers for electrochemical biosensing of carbendazim. Anal Chem 89:3138–3145. https://doi.org/10.1021/acs.analchem.6b04914
Mahdavi V, Ghorbani-Paji F, Ramezani MK, Ghassempour A, Aboul-Enein HY (2019) Dissipation of carbendazim and its metabolites in cucumber using liquid chromatography tandem mass spectrometry. Int J Environ An Ch 99:968–976. https://doi.org/10.1080/03067319.2019.1617281
Strickland AD, Batt CA (2009) Detection of carbendazim by surface-enhanced Raman scattering using cyclodextrin inclusion complexes on gold nanorods. Anal Chem 81:2895–2903. https://doi.org/10.1021/ac801626x
Patel GM, Rohit JV, Singhal RK, Kailasa SK (2015) Recognition of carbendazim fungicide in environmental samples by using 4-aminobenzenethiol functionalized silver nanoparticles as a colorimetric sensor. Sensor Actuat B-Chem 206:684–691. https://doi.org/10.1016/j.snb.2014.09.095
Tu XL, Gao F, Ma X, Zou J, Yu YF, Li MF, Qu FL, Huang XG, Lu LM (2020) Mxene/carbon nanohorn/beta-cyclodextrin-metal-organic frameworks as high-performance electrochemical sensing platform for sensitive detection of carbendazim pesticide. J Hazard Mater 396:122776. https://doi.org/10.1016/j.jhazmat.2020.122776
Zhu CX, Liu D, Chen Z, Li LB, You TY (2019) An ultra-sensitive aptasensor based on carbon nanohorns/gold nanoparticles composites for impedimetric detection of carbendazim at picogram levels. J Colloid Interface Sci 546:92–100. https://doi.org/10.1016/j.jcis.2019.03.035
Gao XY, Gao Y, Bian CC, Ma HY, Liu HL (2019) Electroactive nanoporous gold driven electrochemical sensor for the simultaneous detection of carbendazim and methyl parathion. Electrochim Acta 310:78–85. https://doi.org/10.1016/j.electacta.2019.04.120
Xu GL, Huo DQ, Hou CJ, Zhao YN, Bao J, Yang M, Fa HB (2018) A regenerative and selective electrochemical aptasensor based on copper oxide nanoflowers-single walled carbon nanotubes nanocomposite for chlorpyrifos detection. Talanta 178:1046–1052. https://doi.org/10.1016/j.talanta.2017.08.086
Cui RJ, Xu D, Xie XH, Yi YY, Quan Y, Zhou MX, Gong JJ, Han ZD, Zhang GH (2017) Phosphorus-doped helical carbon nanofibers as enhanced sensing platform for electrochemical detection of carbendazim. Food Chem 221:457–463. https://doi.org/10.1016/j.foodchem.2016.10.094
Santana PCA, Lima JBS, Santana TBS, Santos LFS, Matos CRS, da Costa LP, Gimenez IF, Sussuchi EM (2019) Semiconductor nanocrystals-reduced graphene composites for the electrochemical detection of carbendazim. J Braz Chem Soc 30:1302–1308. https://doi.org/10.21577/0103-5053.20190026
Zhang M, Zhao HT, Yang X, Dong AJ, Zhang H, Wang J, Liu GY, Zhai XC (2016) A simple and sensitive electrochemical sensor for new neonicotinoid insecticide Paichongding in grain samples based on beta-cyclodextrin-graphene modified glassy carbon electrode. Sensors Actuators B Chem 229:190–199. https://doi.org/10.1016/j.snb.2016.01.119
Fei AR, Liu Q, Huan J, Qian J, Dong XY, Qiu BJ, Mao HP, Wang K (2015) Label-free impedimetric aptasensor for detection of femtomole level acetamiprid using gold nanoparticles decorated multiwalled carbon nanotube-reduced graphene oxide nanoribbon composites. Biosens Bioelectron 70:122–129. https://doi.org/10.1016/j.bios.2015.03.028
Li RY, Zhu HY, Li ZJ, Liu JK (2018) Electrochemical determination of acetaminophen using a glassy carbon electrode modified with a hybrid material consisting of graphene aerogel and octadecylamine-functionalized carbon quantum dots. Microchim Acta 185:145. https://doi.org/10.1007/s00604-018-2688-7
Li RY, Yang TT, Li ZJ, Gu ZG, Wang GL, Liu JK (2017) Synthesis of palladium@gold nanoalloys/nitrogen and sulphur-functionalized multiple graphene aerogel for electrochemical detection of dopamine. Anal Chim Acta 954:43–51. https://doi.org/10.1016/j.aca.2016.12.031
Huynh Vu N, Anna G, Min-Ho L (2020) Quantitative determination of triiodothyronine by electrochemical impedance spectroscopic biosensor using gold nanoparticle-modified electrode. J Nanosci Nanotechnol 20:7163–7168. https://doi.org/10.1166/jnn.2020.18853
Lee HE, Ahn HY, Mun J, Lee YY, Kim M, Cho NH, Chang K, Kim WS, Rho J, Nam KT (2018) Amino-acid- and peptide-directed synthesis of chiral plasmonic gold nanoparticles. Nature 556:360–365. https://doi.org/10.1038/s41586-018-0034-1
Smith DK, Korgel BA (2008) The importance of the CTAB surfactant on the colloidal seed-mediated synthesis of gold nanorods. Langmuir 24:644–649. https://doi.org/10.1021/la703625a
Zhang HB, Lu YG, Guan CL, Song N, Zhang YY, Liu H, Fang JZ (2017) One-pot controllable synthesis of the missing uniform icosidodecahedral gold nanocrystals with high-index facets and their plasmonic properties. J Mater Chem C 5:645–653. https://doi.org/10.1039/c6tc04744k
Su LT, Wang S, Wang LM, Yan ZY, Yi HY, Zhang DW, Shen GQ, Ma Y (2020) Fluorescent aptasensor for carbendazim detection in aqueous samples based on gold nanoparticles quenching Rhodamine B. Spectrochim Acta A 225:117511. https://doi.org/10.1016/j.saa.2019.117511
Li LD, Peng Y, Yue YH, Hu Y, Liang X, Yin PG, Guo L (2015) Synthesis of concave gold nanocuboids with high-index facets and their enhanced catalytic activity. Chem Commun 51:11591–11594. https://doi.org/10.1039/c5cc02106e
Xie Y, Gao F, Tu XL, Ma X, Dai RY, Peng GY, Yu YF, Lu LM (2019) Flake-like neodymium molybdate wrapped with multi-walled carbon nanotubes as an effective electrode material for sensitive electrochemical detection of carbendazim. J Electroanal Chem 855:113468. https://doi.org/10.1016/j.jelechem.2019.113468
Li D, Li DW, Li Y, Fossey JS, Long YT (2010) Cyclic electroplating and stripping of silver on Au@SiO2 core/shell nanoparticles for sensitive and recyclable substrate of surface-enhanced Raman scattering. J Mater Chem 20:3688–3693. https://doi.org/10.1039/b924865j
Yang YX, Huo DQ, Wu HX, Wang XF, Yang JS, Bian MH, Ma Y, Hou CJ (2018) P-doped carbon quantum dots as a fluorescent sensing platform for carbendazim detection based on fluorescence resonance energy transfer. Sensor Actuat B-Chem 274:296–303. https://doi.org/10.1016/j.snb.2018.07.130
Tian CH, Zhang SF, Wang HB, Chen C, Han ZD, Chen ML, Zhu YY, Cui RJ, Zhang GH (2019) Three-dimensional nanoporous copper and reduced graphene oxide composites as enhanced sensing platform for electrochemical detection of carbendazim. J Electroanal Chem 847:113243. https://doi.org/10.1016/j.jelechem.2019.113243
Acknowledgements
The authors acknowledge the financial support from The National Key Research and Development Program of China (No. 2018YFC1603001); emergency response to COVID-19 washing products of the Key Laboratory of Synthetic and Biological Colloids, Ministry of Education (No. YJ2020-02); and the National Natural Science Foundation of China (No. 21576115).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 36786 kb)
Rights and permissions
About this article
Cite this article
Jin, W., Ruiyi, L., Nana, L. et al. Electrochemical detection of carbendazim with mulberry fruit-like gold nanocrystal/multiple graphene aerogel and DNA cycle amplification. Microchim Acta 188, 284 (2021). https://doi.org/10.1007/s00604-021-04886-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-021-04886-y