Abstract
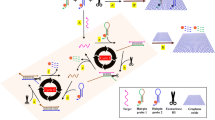
The base-excision repair enzyme uracil-DNA glycosylase (UDG) plays a crucial role in the maintenance of genome integrity. The authors describe a fluorometric method for the detection of the activity of UDG. It is making use of (a) a 3’-FAM-labeled hairpin DNA probe with two uracil deoxyribonucleotides in the self-complementary duplex region of its hairpin structure, (b) exonuclease I (Exo I) that catalyzes the release of FAM from the UDG-induced stretched ssDNA probe, and (c) graphene oxide that quenches the green FAM fluorescence of the intact hairpin DNA probe in the absence of UDG. If Exo I causes the release of FAM from the hairpin DNA probe, the fluorescence peaking at 517 nm is turned off in the absence of UDG but turned on in its presence. The resulting assay has a wide linear range (0.008 to 1 U·mL−1) and a detection limit as low as 0.005 U·mL−1. It has good specificity for UDG over potentially interfering enzymes and gave satisfactory results when applied to biological samples. Conceivably, the method may be used in a wide range of applications such as in diagnosis, drug screening, and in studying the repair of DNA lesions.

Schematic presentation of a fluorometric strategy for detection of the activity of uracil-DNA glycosylase by using on graphene oxide and exonuclease I assisted signal amplification.





Similar content being viewed by others
References
Lu YJ, Hu DP, Deng Q, Wang ZY, Huang BH, Fang YX, Zhang K, Wong WL (2015) Sensitive and selective detection of uracil-DNA glycosylase activity with a new pyridinium luminescent switch-on molecular probe. Analyst 140:5998–6004. https://doi.org/10.1039/c5an01158b
Wu YS, Wang L, Zhu J, Jiang W (2015) A DNA machine-based fluorescence amplification strategy for sensitive detection of uracil-DNA glycosylase activity. Biosens Bioelectron 68:654–659. https://doi.org/10.1016/j.bios.2015.01.069
Ma Y, Zhao J, Li X, Zhang L, Zhao S (2015) A label free fluorescent assay for uracil-DNA glycosylase activity based on the signal amplification of exonuclease I. RSC Adv 5:80871–80874. https://doi.org/10.1039/c5ra12958c
Khusbu FY, Zhou X, Chen H, Ma C, Wang K (2018) Thioflavin T as a fluorescence probe for biosensing applications. Trends Anal Chem 109:1–18. https://doi.org/10.1016/j.trac.2018.09.013
Wang LJ, Ren M, Zhang QY, Tang B, Zhang CY (2017) Excision repair-initiated enzyme-assisted bicyclic cascade signal amplification for ultrasensitive detection of uracil-DNA glycosylase. Anal Chem 89:4488–4494. https://doi.org/10.1021/acs.analchem.6b04673
Kruman II, Schwartz E, Kruman Y, Cutler RG, Zhu X, Greig NH, Mattson MP (2004) Suppression of uracil-DNA glycosylase induces neuronal apoptosis. J Biol Chem 279:43952–43960. https://doi.org/10.1074/jbc.M408025200
Chen L, Long Y, Liu B, Xiang D, Zhu H (2014) Real time monitoring uracil excision using uracil-containing molecular beacons. Anal Chim Acta 819:71–77. https://doi.org/10.1016/j.aca.2014.02.002
Zhang Y, Li CC, Tang B, Zhang CY (2017) Homogeneously sensitive detection of multiple DNA glycosylases with intrinsically fluorescent nucleotides. Anal Chem 89:7684–7692. https://doi.org/10.1021/acs.analchem.7b01655
Imai K, Slupphaug G, Lee WI, Revy P, Nonoyama S, Catalan N, Yel L, Forveille M, Kavli B, Krokan HE, Ochs HD, Fischer A, Durandy A (2003) Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol 4:1023–1028. https://doi.org/10.1038/ni974
Sousa MM, Krokan HE, Slupphaug G (2007) DNA-uracil and human pathology. Mol Asp Med 28:276–273. https://doi.org/10.1016/j.mam.2007.04.006
Hu D, Huang Z, Pu F, Ren J, Qu X (2011) A label-free, Quadruplex-based functional molecular beacon (LFG4-MB) for fluorescence turn-on detection of DNA and nuclease. Chem Eur J 17:1635–1641. https://doi.org/10.1002/chem.201001331
Wu K, Ma C, Deng Z, Fang N, Tang Z, Zhu X, Wang K (2018) Label-free and nicking enzyme-assisted fluorescence signal amplification for RNase H analysis based on a G-quadruplexe/thioflavin T complex. 182: 142–147. doi: https://doi.org/10.1016/j.talanta.2018.01.075
Du YC, Cui YX, Li XY, Sun GY, Zhang YP, Tang AN, Kim K, Kong DM (2018) Terminal deoxynucleotidyl transferase and T7 exonuclease-aided amplification strategy for ultrasensitive detection of uracil-DNA glycosylase. Anal Chem 90:8629–8634. https://doi.org/10.1021/acs.analchem.8b01928
Prorok P, Alili D, Saint-Pierre C, Gasparutto D, Zharkov DO, Ishchenko AA, Tudek B, Saparbaev MK (2015) Uracil in duplex DNA is a substrate for the nucleotide incision repair pathway in human cells. Proceedings Natl. Acad Sci U S A 110:3695–3703. https://doi.org/10.1073/pnas.1305624110
Liu X, Chen M, Hou T, Wang X, Liu S, Li F (2013) A novel electrochemical biosensor for label-free detection of uracil DNA glycosylase activity based on enzyme-catalyzed removal of uracil bases inducing strand release. Electrochim Acta 113:514–518. https://doi.org/10.1016/j.electacta.2013.09.131
Jiao F, Qian P, Qin Y, Xia Y, Deng C, Nie Z (2016) A novel and label-free biosensors for uracil-DNA glycosylase activity based on the electrochemical oxidation of guanine bases at the graphene modified electrode. Talanta 147:98–102. https://doi.org/10.1016/j.talanta.2015.09.045
Nie H, Wang W, Li W, Nie Z, Yao S (2015) A colorimetric and smartphone readable method for uracil-DNA glycosylase detection based on the target-triggered formation of G-quadruplex. Analyst 140:2771–2777. https://doi.org/10.1039/c4an02339k
Nguyen V, Le D, Nie C, Zhou D, Wang Y, Tang L, Jiang J, Yu R (2012) Enzyme-catalyzed assembly of gold nanoparticles for visualized screening of DNA base excision repair. Talanta 100:303–307. https://doi.org/10.1016/j.talanta.2012.07.065
Tao J, Song PS, Sato Y, Nishizawa S, Teramae N, Tong A, Yu Xiang Y (2015) A label-free and sensitive fluorescent method for the detection of uracil-DNA glycosylase activity. Chem Commun 51:929–932. https://doi.org/10.1039/c4cc06170e
Liu XJ, Che MQ, Hou T, Wang XZ, Liu SF, Li F (2014) Label-free colorimetric assay for base excision repair enzyme activity based on nicking enzyme assisted signal amplification. Biosens Bioelectron 54:598–602. https://doi.org/10.1016/j.bios.2013.11.062
Ma CB, Wu KF, Liu HS, Xia K, Wang KM, Wang J (2016) Label-free fluorescence turn-on detection of uracil DNA glycosylase activity based on G-quadruplex formation. Talanta 160:449–453. https://doi.org/10.1016/j.talanta.2016.07.048
Ahn JK, Lee CY, Park KS, Park HG (2018) Abasic site-assisted inhibition of nicking endonuclease activity for the sensitive determination of uracil DNA glycosylase. Biotechnol J 13:170603. https://doi.org/10.1002/biot.201700603
Wu YS, Wang L, Jiang W (2017) Toehold-mediated strand displacement reaction-dependent fluorescent strategy for sensitive detection of uracil-DNA glycosylase activity. Biosens Bioelectron 89:984–988. https://doi.org/10.1016/j.bios.2016.10.053
Wu YS, Yan P, Xu XW, Jiang W A unique dual recognition hairpin probe mediated fluorescence amplification method for sensitive detection of uracil-DNA glycosylase and endonuclease IV activities. Analyst 141:1789–1795. https://doi.org/10.1039/c5an02483h
Liu DK, Lu X, Yang YW, Zhai YY, Zhang J, Li L (2018) A novel fluorescent aptasensor for the highly sensitive and selective detection of cardiac troponin I based on a graphene oxide platform. Anal Bioanal Chem 410:4285–4291. https://doi.org/10.1007/s00216-018-1076-9
Xiao KY, Liu J, Chen H, Zhang S, Kong JL (2017) A label-free and high-efficient GO-based aptasensor for cancer cells based on cyclic enzymatic signal amplification. Biosens Bioelectron 91:76–81. https://doi.org/10.1016/j.bios.2016.11.057
Li MK, Hu LY, Niu CG, Huang DW, Zeng GM (2018) A fluorescent DNA based probe for hg(II) based on thymine-hg(II)-thymine interaction and enrichment via magnetized graphene oxide. Microchim Acta 185:207. https://doi.org/10.1007/s00604-018-2689-6
Ma CB, Wu KF, Zhao H, Liu HS, Wang KM, Xia K (2018) Fluorometric aptamer-based determination of ochratoxin a based on the use of graphene oxide and RNase H-aided amplification. Microchim Acta 185:347. https://doi.org/10.1007/s00604-018-2885-4
Zhang H, Zhang H, Aldalbahi A, Zuo XL, Fan CH, Mi XQ (2017) Fluorescent biosensors enabled by graphene and graphene oxide. Biosens Bioelectron 89:96–106. https://doi.org/10.1016/j.bios.2016.07.030
Li CH, Xiao X, Tao J, Wang DM, Huang CZ, Zhen SJ (2017) A graphene oxide-based strand displacement amplification platform for ricin detection using aptamer as recognition element. Biosens Bioelectron 91:149–154. https://doi.org/10.1016/j.bios.2016.12.010
Chen J, Ge J, Zhang L, Li ZH, Li JJ, Sun YJ, Qu LB (2016) Reduced graphene oxide nanosheets functionalized with poly (styrene sulfonate) as a peroxidase mimetic in a colorimetric assay for ascorbic acid. Microchim Acta 183:1847–1853. https://doi.org/10.1007/s00604-016-1826-3
Wu KF, Ma C, Zhao H, Chen M, Deng Z (2019) Sensitive aptamer-based fluorescene assay for ochratoxin a based on RNase H signal amplification. Food Chem 277:273–278. https://doi.org/10.1016/j.foodchem.2018.10.130
Sun Y, Peng P, Guo R, Wang H, Li T (2018) Exonuclease III-boosted cascade reactions for ultrasensitive SERS detection of nucleic acids. Biosens Bioelectron 104:32–38. https://doi.org/10.1016/j.bios.2017.12.047
Wu T, Yang Y, Chen W, Wang J, Yang Z, Wang S, Xiao X, Li M, Zhao M (2018) Noncanonical substrate preference of lambda exonuclease for 5′-nonphosphate-ended dsDNA and a mismatch-induced acceleration effect on the enzymatic reaction. Nucleic Acids Res 46:3119–3129. https://doi.org/10.1093/nar/gky154
Dong JJ, Lian JY, Jin Y, Baoxin Li BX (2017) Guanine-based chemiluminescence resonance energy transfer biosensing platform for the specific assay of uracil-DNA glycosylase activity. Anal Methods 9:276–281. https://doi.org/10.1039/c6ay02964g
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 21205142, 31370104), State Key Laboratory of Chemo/Biosensing and Chemometrics, Hunan University (2017006), The Research Innovation Program for Graduates of Central South University (2018zzts384, 2018zzts399).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author(s) declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 396 kb)
Rights and permissions
About this article
Cite this article
Chen, M., Li, W., Ma, C. et al. Fluorometric determination of the activity of uracil-DNA glycosylase by using graphene oxide and exonuclease I assisted signal amplification. Microchim Acta 186, 110 (2019). https://doi.org/10.1007/s00604-019-3247-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3247-6