Abstract
The authors describe an antibody-aptamer based hetero-sandwich amperometric biosensor for the foodborne pathogen Vibrio parahaemolyticus. Antibody on the surface of a gold electrode first captures the target pathogen, and then the immunocomplex is incubated with an aptamer consisting of an ssDNA probe. A hetero-sandwich structure is formed which allows for signal enhancement by rolling circle amplification. Following addition of Methylene Blue as an electrochemical DNA probe, the amperometric signal, best measured at 0.28 V (vs. Ag/AgCl), covers the 2.2 to 2.2*108 cfu per mL concentration range, and the limit of detection is as low as 2 cfu per mL. The method was successfully applied to the quantification of V. parahaemolyticus in spiked fish samples. In our perception, this hetero-sandwich electrochemical biosensor provides an ultrasensitive tool for the quantitation of a variety of pathogen if appropriate antibodies and aptamers are available.

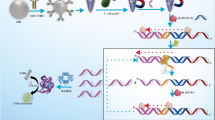
Schematic image of signal amplified hetero-sandwich electrochemical biosensor for Vibrio parahaemolyticus detection. Rolling circular amplification was adopted as the signal amplification strategy on the hetero-sandwich electrochemical biosensor for sensitive and accurate detection of Vibrio parahaemolyticus by simultaneous using of antibody and aptamer as the recognition probes. Excellent improvement of sensitivity and satisfied results were achieved for Vibrio parahaemolyticus in food samples.




Similar content being viewed by others
References
Duan N, Wu S, Ma X, Yu X, Wang Z (2014) A universal fluorescent aptasensor based on AccuBlue dye for the detection of pathogenic bacteria. Anal Biochem 454:1–6
Suffredini E, Mioni R, Mazzette R, Bordin P, Serratore P, Fois F, Piano A, Cozzi L, Croci L (2014) Detection and quantification of Vibrio parahaemolyticus in shellfish from Italian production areas. Int J Food Microbiol 184:14–20
Sakata J, Kawatsu K, Iwasaki T, Kumeda Y (2015) Development of a rapid and simple immunochromatographic assay to identify Vibrio parahaemolyticus. J Microbiol Methods 116:23–29
Raghunath P, Acharya S, Bhanumathi A, Karunasagar I, Karunasagar I (2008) Detection and molecular characterization of Vibrio parahaemolyticus isolated from seafood harvested along the southwest coast of India. Food Microbiol 25:824–830
Garrido-Maestu A, Chapela MJ, Peñaranda E, Vieites JM, Cabado AG (2014) In-house validation of novel multiplex real-time PCR gene combination for the simultaneous detection of the main human pathogenic vibrios. Food Control 37:371–379
Rizvi AV, Bej AK (2010) Multiplexed real-time PCR amplification of tlh, tdh and trh genes in Vibrio parahaemolyticus and its rapid detection in shellfish and Gulf of Mexico water. Antonie Van Leeuwenhoek 98:279–290
Pechorsky A, Nitzan Y, Lazarovitch T (2009) Identification of pathogenic bacteria in blood cultures: comparison between conventional and PCR methods. J Microbiol Methods 78:325–330
Juvonen R, Koivula T, Haikara A (2008) Group-specific PCR-RFLP and real-time PCR methods for detection and tentative discrimination of strictly anaerobic beer-spoilage bacteria of the class clostridia. Int J Food Microbiol 125:162–169
Raghunath P, Karunasagar I, Karunasagar I (2009) Improved isolation and detection of pathogenic Vibrio parahaemolyticus from seafood using a new enrichment broth. Int J Food Microbiol 129:200–203
Santiago-Felipe S, Tortajada-Genaro LA, Puchades R, Maquieira A (2014) Recombinase polymerase and enzyme-linked immunosorbent assay as a DNA amplification-detection strategy for food analysis. Anal Chim Acta 811:81–87
Hamula CLA, Zhang HQ, Guan LL, Li XF, Le XC (2008) Selection of aptamers against live bacterial cells. Anal Chem 80:7812–7819
Ellington AD, Szostak JW (1990) In vitro selection RNA molecules bind specific ligands. Nature 346:818–822
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510
Teng J, Yuan F, Ye YW, Zheng L, Yao L, Xue F, Chen W, Li BG (2016) Aptamer-based Technologies in Foodborne Pathogen Detection. Front Microbiol 7(1426):1–11
Wu JJ, Zhu YY, Xue F, Mei ZL, Yao L, Wang X, Zheng L, Liu J, Liu GD, Peng CF, Chen W (2014) Recent trends in SELEX technique and its application to food safety monitoring. Microchim Acta 181:479–491
Yoo SM, Kim DK, Lee SY (2015) Aptamer-functionalized localized surface plasmon resonance sensor for the multiplexed detection of different bacterial species. Talanta 132:112–117
Wu W, Li J, Pan D, Li J, Song S, Rong M, Li Z, Gao J, Lu J (2014) Gold nanoparticle-based enzyme-linked antibody-aptamer sandwich assay for detection of Salmonella typhimurium. ACS Appl Mater Interfaces 6:16974–16981
Wu S, Duan N, Shi Z, Fang C, Wang Z (2014) Simultaneous aptasensor for multiplex pathogenic bacteria detection based on multicolor upconversion nanoparticles labels. Anal Chem 86:3100–3107
Ma X, Jiang Y, Jia F, Yu Y, Chen J, Wang Z (2014) An aptamer-based electrochemical biosensor for the detection of Salmonella. J Microbiol Methods 98:94–98
Xie Y, Xu L, Wang Y, Shao J, Wang L, Wang H, Qian H, Yao W (2013) Label-free detection of the foodborne pathogens of Enterobacteriaceae by surface-enhanced Raman spectroscopy. Anal Methods 5:946–952
Paniel N, Baudart J, Hayat A, Barthelmebs L (2013) Aptasensor and genousensor methods for detection of microbes in real world samples. Methods 64:229–240
Donolato M, Antunes P, Torre TZ, Hwu ET, Chen CH, Burger R, Rizzi G, Bosco FG, Stromme M, Boisen A, Hansen MF (2015) Quantification of rolling circle amplified DNA using magnetic nanobeads and a Blu-ray optical pick-up unit. Biosens Bioelectron 67:649–655
Huang L, Wu JJ, Zheng L, Qian HS, Xue F, Wu YC, Pan DD, Adeloju SB, Chen W (2013) Rolling chain amplification based signal-enhanced electrochemical aptasensor for ultrasensitive detection of ochratoxin a. Anal Chem 85:10842–10849
Zhou L, LJ O, Chu X, Shen GL, Yu RQ (2007) Aptamer-based rolling circle amplification: a platform for electrochemical detection of protein. Anal Chem 29:7492–7500
Bi S, Ji B, Zhang Z, Zhang S (2013) A chemiluminescence imaging array for the detection of cancer cells by dual-aptamer recognition and bio-bar-code nanoprobe-based rolling circle amplification. Chem Commun 49:3452–3454
Pincella F, Song Y, Ochiai T, Isozaki K, Sakamoto K, Miki K (2014) Square-centimeter-scale 2D-arrays of au@ag core–shell nanoparticles towards practical SERS substrates with enhancement factor of 107. Chem Phys Lett 605-606:115–120
Mei ZL, Qu W, Deng Y, Chu HQ, Cao JX, Xue F, Zheng L, El-Nezamic HS, Wu YC, Chen W (2013) One-step signal amplified lateral flow strip biosensor for ultrasensitive and on-site detection of bisphenol a (BPA) in aqueous samples. Biosens Bioelectron 49:457–461
Liu K, Yan X, Mao B, Wang S, Deng L (2016) Aptamer-based detection of Salmonella enteritidis using double signal amplification by Klenow fragment and dual fluorescence. Microchim Acta 183:63–649
Lan MB, Zhou Q, Zhao YH, Teng YJ, Chen C, Zhao HL, Yuan HH (2010) Electrochemical detection of a Vibrio parahaemolyticus sequence-specific gene based on a gold electrode modified with a single stranded probe. Sci China: Chem 53:1366–1370
Zhao GY, Xing FF, Deng SP (2007) A disposable amperometric enzyme immunosensor for rapid detection of Vibrio parahaemolyticus in food based on agarose/Nano-au membrane and screen-printed electrode Electrochem. Commun 9:1263–1268
Wang XY, Sun DP, Tong YL, Zhong YS, Chen ZG (2017) A voltammetric aptamer-based thrombin biosensor exploiting signal amplification via synergetic catalysis by DNAzyme and enzyme decorated AuPd nanoparticles on a poly(o-phenylenediamine) support. Microchim Acta 184:1791–1799
Rafati A, Gill P (2015) Microfluidic method for rapid turbidimetric detection of the DNA of mycobacterium tuberculosis using loop-mediated isothermal amplification in capillary tubes. Microchim Acta 182(3–4):523–530
Liu S, Leng X, Wang X, Pei Q, Cui X, Wang Y, Huang J (2017) Enzyme-free colorimetric assay for mercury (II) using DNA conjugated to gold nanoparticles and strand displacement amplification. Microchim Acta. doi:10.1007/s00604-017-2182-7
Wang X, Liu W, Yin B, Sang Y, Liu Z, Dai Y, Tao Z (2017) An isothermal strand displacement amplification strategy for nucleic acids using junction forming probes and colorimetric detection. Microchim Acta 184:1603–1610
Yamanaka ES, Tortajada-Genaro LA, Maquieira Á (2017) Low-cost genotyping method based on allele-specific recombinase polymerase amplification and colorimetric microarray detection. Microchim Acta. doi:10.1007/s00604-017-2144-0
Acknowledgements
This work is financially supported by the Science and Technology Ministry of China (2015BAD17B02-3), the NSFC Grant of 21475030, the S&T Research Project of Anhui Province 15czz03109, the National 10000 Talents-Youth Top-notch Talent Program, and the KC Wong Magna Fund in Ningbo University.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The author(s) declare that they have no competing interests.
Electronic supplementary material
ESM 1
(DOCX 540 kb)
Rights and permissions
About this article
Cite this article
Teng, J., Ye, Y., Yao, L. et al. Rolling circle amplification based amperometric aptamer/immuno hybrid biosensor for ultrasensitive detection of Vibrio parahaemolyticus . Microchim Acta 184, 3477–3485 (2017). https://doi.org/10.1007/s00604-017-2383-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-017-2383-0