Abstract
Purpose
Tissue disaggregation and the cell sorting technique by surface markers has played an important role in isolating lymphatic endothelial cells (LECs) from lymphatic malformation (LM). However, this technique may have the drawback of impurities or result in isolation failure because it is dependent on surface marker expressions, the heterogeneity of which has been found in the lymphatic system. We developed a novel method for isolating LM-LECs without using whole tissue disaggregation.
Methods
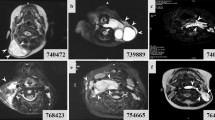
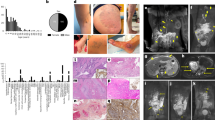
Seven LM surgical specimens were collected from seven patients with LMs. LM-LECs were detached from the LM cyst wall by “lumen digestion” and irrigating the cystic cavity with trypsin, and maintained in culture.
Results
The cells formed a monolayer with a cobblestone-like appearance. Immunohistochemistry and quantitative RT-PCR of these cells revealed high expression of lymphatic-specific genes, confirming their identity as LM-LECs. The whole-exome sequencing and PIK3CA sequencing of these cells revealed somatic mutations in PIK3CA in all cases.
Conclusions
We established a novel technique for isolating LM-LECs from LM tissue by “lumen digestion” without whole-tissue disaggregation. The limited incorporation of non-LM LECs in the isolate in our method could make it an important tool for investigating the heterogeneity of gene expression as well as mutations in LM-LECs.




Similar content being viewed by others
References
Florez-Vargas A, Vargas SO, Debelenko LV, Perez-Atayde AR, Archibald T, Kozakewich HP, et al. Comparative analysis of D2–40 and LYVE-1 immunostaining in lymphatic malformations. Lymphology. 2008;41:103–10.
Wassef M, Blei F, Adams D, Alomari A, Baselga E, Berenstein A, et al. ISSVA Board and Scientific Committee. Vascular anomalies classification: recommendations from the international society for the study of vascular anomalies. Pediatrics. 2015;136:e203–e214214.
Schoinohoriti OK, Theologie-Lygidakis N, Tzerbos F, Iatrou I. Lymphatic malformations in children and adolescents. J Craniofac Surg. 2012;23:1744–7.
Huang HY, Ho CC, Huang PH, Hsu SM. Co-expression of VEGF-C and its receptors, VEGFR-2 and VEGFR-3, in endothelial cells of lymphangioma. Implication in autocrine or paracrine regulation of lymphangioma. Lab Invest. 2001;81:1729–34.
Osborn AJ, Dickie P, Neilson DE, Glaser K, Lynch KA, Gupta A, et al. Activating PIK3CA alleles and lymphangiogenic phenotype of lymphatic endothelial cells isolated from lymphatic malformations. Hum Mol Genet. 2015;24:926–38.
Luks VL, Kamitaki N, Vivero MP, Uller W, Rab R, Bovee JV, et al. Lymphatic and other vascular malformative/overgrowth disorders are caused by somatic mutations in PIK3CA. J Pediatr. 2015;166:1048–54.
Padia R, Zenner K, Bly R, Bennett J, Bull C, Perkins J. Clinical application of molecular genetics in lymphatic malformations. Laryngoscope Investig Otolaryngol. 2019;4:170–3.
Blesinger H, Kaulfuß S, Aung T, Schwoch S, Prantl L, Rößler J, et al. PIK3CA mutations are specifically localized to lymphatic endothelial cells of lymphatic malformations. ProS One. 2018;7:e0200343.
Lokmic Z, Mitchell GM, Chong NKW, Bastiaanse J, Gerrand YW, Zeng Y, et al. Isolation of human lymphatic malformation endothelial cells, their in vitro characterization and in vivo survival in a mouse xenograft model. Angiogenesis. 2014;17:1–15.
Kaipainen A, Chen E, Chang L, Zhao B, Shin H, Stahl A, et al. Characterization of lymphatic malformations using primary cells and tissue transcriptomes. Scand J Immunol. 2019;90:e12800.
Tsurusaki Y, Ohashi I, Enomoto Y, Naruto T, Mitsui J, Aida N, et al. A novel UBE2A mutation causes X-linked intellectual disability type Nascimento. Hum Genome Var. 2017;4:17019.
Kawai Y, Minami T, Fujimori M, Hosaka K, Mizuno R, Ikomi F, et al. Characterization and microarray analysis of genes in human lymphatic endothelial cells from patients with breast cancer. Lymphat Res Biol. 2007;5:115–26.
Kawai Y, Hosaka K, Kaidoh M, Minami T, Kodama T, Ohhashi T. Heterogeneity in immunohistochemical, genomic, and biological properties of human lymphatic endothelial cells between initial and collecting lymph vessels. Lymphat Res Biol. 2008;6:15–27.
Bittinger F, Klein CL, Skarke C, Brochhausen C, Walgenbach S, Röhrig O, et al. PECAM-1 expression in human mesothelial cells: an in vitro study. Pathobiology. 1996;64:320–7.
Ordóñez NG. D2–40 and podoplanin are highly specific and sensitive immunohistochemical markers of epithelioid malignant mesothelioma. Hum Pathol. 2005;36:372–80.
Galambos C, Nodit L. Identification of lymphatic endothelium in pediatric vascular tumors and malformations. Pediatr Dev Pathol. 2005;8:181–9.
Norgall S, Papoutsi M, Rössler J, Schweigerer L, Wilting J, Weich HA. Elevated expression of VEGFR-3 in lymphatic endothelial cells from lymphangiomas. BMC Cancer. 2007;7:105.
Boscolo E, Coma S, Luks VL, Greene AK, Klagsbrun M, Warman ML, et al. AKT hyper-phosphorylation associated with PI3K mutations in lymphatic endothelial cells from a patient with lymphatic malformation. Angiogenesis. 2015;18:151–62.
Gymnopoulos M, Elsliger MA, Vogt PK. Rare cancer-specific mutations in PIK3CA show gain of function. Proc Natl Acad Sci USA. 2007;104:5569–74.
Keppler-Noreuil KM, Rios JJ, Parker VE, Semple RK, Lindhurst MJ, Sapp JC, et al. PIK3CA-related overgrowth spectrum (PROS): diagnostic and testing eligibility criteria, differential diagnosis, and evaluation. Am J Med Genet A. 2015;167A:287–95.
Mirzaa G, Timms AE, Conti V, Boyle EA, Girisha KM, Martin B, et al. PIK3CA-associated developmental disorders exhibit distinct classes of mutations with variable expression and tissue distribution. JCI Insight. 2016;1:e87623.
Yeung KS, Ip JJ, Chow CP, Kuong EY, Tam PK, Chan GC, et al. Somatic PIK3CA mutations in seven patients with PIK3CA-related overgrowth spectrum. Am J Med Genet A. 2017;173:978–84.
Wiegand S, Wichmann G, Dietz A. Treatment of lymphatic malformations with the mTOR inhibitor sirolimus: a systematic review. Lymphat Res Biol. 2018;16:330–9.
Funding
This study was funded by the Kanagawa Prefectural Hospital’s Child Health Care and Research Foundation (to H.U.), the Naito Foundation (to Y.T.), and the Yokohama Foundation for Advancement of Medical Science (to Y.T.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Usui, H., Tsurusaki, Y., Shimbo, H. et al. A novel method for isolating lymphatic endothelial cells from lymphatic malformations and detecting PIK3CA somatic mutation in these isolated cells. Surg Today 51, 439–446 (2021). https://doi.org/10.1007/s00595-020-02122-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00595-020-02122-3