Abstract
Aims
Diabetic nephropathy (DN) is a diabetes-related chronic vasculitis. DN diminishes kidney function over time and, of course, leads to end stage renal disease in people (ESRD). In spite of the advances in diagnostic and treatment methods for DN, DN continues to impose a significant physical and psychological burden on patients, severely impacting their quality of life, making the hunt for novel therapeutic targets necessary.
Methods
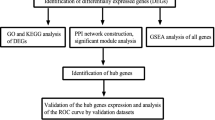
The Gene Expression Omnibus (GEO) microarray datasets GSE1009, GSE30122, GSE142153, and GSE96804 were downloaded to identify differentially expressed genes (DEGs) in kidney tissues from patients in the DN group and normal controls. These three datasets were examined for genes associated with basement membranes (BMs) with differential gene expression. The target genes were then subjected to gene ontology (GO) annotation and Kyoto Gene and Genome Encyclopedia (KEGG) pathway enrichment analysis. BM-related genes underwent PPI network analysis and screening of the top 10 hub genes, along with immune infiltration analysis and column line graph model development. Finally, we conducted DN therapeutic medication prediction and the creation of something like a miRNA network for genetic markers with BMs.
Results
Seven candidate BM-related genes (COL4A1, COL4A2, COL6A2, COL6A3, FN1, ITGQ4, and LAMB1) with acceptable helps the healthcare were discovered. Enrichment analysis of diabetes-related genes event occurred the role of biological processes including extracellular matrix organization, extracellular structural organization, and collagen-containing extracellular matrix, as well as the PI3K-Akt signaling pathway and the AGE-RAGE signaling pathway, in diabetic complications. These genes may also be associated in immune cells and autoimmune activities, such as Macrophages and MHC class I, in order to impact the immune process in DN. In the meanwhile, based on these seven BM-related genes, we discovered that Ginsenoside Rh1 was very significant for drug targeting.
Conclusions
This research identified seven BM-related genes as possible diagnostic and therapeutic biomarkers for DN. Analysis of inflammatory infiltration indicated that these genes may be important in inflammatory processes through Macrophages and MHC class I, hence impacting the course and development of DN illness. The development of a correlated column line graph model for it also shown excellent predictive capabilities. In addition, we have found pharmaceuticals, such as Ginsenoside Rh1, that may provide fresh insights into the personalized management of patients with DN.










Similar content being viewed by others
Data availability
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
References
Molinaro R, Dauscher C (2017) Complications resulting from uncontrolled diabetes. MLO Med Lab Obs 49(2):20–22
Gross JL, de Azevedo MJ, Silveiro SP et al (2005) Diabetic nephropathy: diagnosis, prevention, and treatment. Diabetes Care 28(1):164–176
Rheinberger M, Böger CA (2014) Diabetische nephropathie: neues in diagnose, prävention und therapie diabetic nephropathy: new insights into diagnosis, prevention and treatment. Dtsch Med Wochenschr 139(14):704–706
Xiang E, Han B, Zhang Q et al (2020) Human umbilical cord-derived mesenchymal stem cells prevent the progression of early diabetic nephropathy through inhibiting inflammation and fibrosis. Stem Cell Res Ther 11(1):336
Garud MS, Kulkarni YA (2014) Hyperglycemia to nephropathy via transforming growth factor beta. Curr Diabetes Rev 10(3):182–189
Afkarian M, Zelnick LR, Hall YN et al (2016) Clinical manifestations of kidney disease among us adults with diabetes, 1988–2014. JAMA 316(6):602–610
Loganathan TS, Sulaiman SA, Abdul Murad NA et al (2020) Interactions among non-coding rnas in diabetic nephropathy. Front Pharmacol 11:191
Rossing P, Persson F, Frimodt-Møller M (2018) Prognosis and treatment of diabetic nephropathy: Recent advances and perspectives. Nephrol Ther 14(Suppl 1):S31–S37
Ogurtsova K, da Rocha Fernandes JD, Huang Y et al (2017) IDF diabetes atlas: global estimates for the prevalence of diabetes for 2015 and 2040. Diabetes Res Clin Pract 128:40–50
Sekiguchi R, Yamada KM (2018) Basement membranes in development and disease. Curr Top Dev Biol 130:143–191
Yurchenco PD (2011) Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb Perspect Biol 3(2):a004911
Pozzi A, Yurchenco PD, Iozzo RV (2017) The nature and biology of basement membranes. Matrix Biol 57–58:1–11
Jayadev R, Chi Q, Keeley DP et al (2019) α-Integrins dictate distinct modes of type IV collagen recruitment to basement membranes. J Cell Biol 218(9):3098–3116
Li S, Qi Y, McKee K et al (2017) Integrin and dystroglycan compensate each other to mediate laminin-dependent basement membrane assembly and epiblast polarization. Matrix Biol 57–58:272–284
Foster MH (2017) Basement membranes and autoimmune diseases. Matrix Biol 57–58:149–168
Randles M, Lausecker F, Kong Q et al (2021) Identification of an altered matrix signature in kidney aging and disease. J Am Soc Nephrol 32(7):1713–1732
Baelde HJ, Eikmans M, Doran PP et al (2004) Gene expression profiling in glomeruli from human kidneys with diabetic nephropathy. Am J Kidney Dis 43(4):636–650
Woroniecka KI, Park AS, Mohtat D et al (2011) Transcriptome analysis of human diabetic kidney disease. Diabetes 60(9):2354–2369
Sur S, Nguyen M, Boada P et al (2021) FcER1: a novel molecule implicated in the progression of human diabetic kidney disease. Front Immunol 12:769972
Pan Y, Jiang S, Hou Q et al (2018) Dissection of glomerular transcriptional profile in patients with diabetic nephropathy: srgap2a protects podocyte structure and function. Diabetes 67(4):717–730
Jayadev R, Morais MRPT, Ellingford JM et al (2022) A basement membrane discovery pipeline uncovers network complexity, regulators, and human disease associations. Sci Adv 8(20):eabn2265
Szklarczyk D, Gable AL, Nastou KC et al (2021) The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 49(D1):D605–D612
Doncheva NT, Morris JH, Gorodkin J et al (2019) Cytoscape StringApp: network analysis and visualization of proteomics data. J Proteome Res 18(2):623–632
Rooney MS, Shukla SA, Wu CJ et al (2015) Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160(1–2):48–61
Kuleshov MV, Jones MR, Rouillard AD et al (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44(W1):W90–W97
Pannebakker MA, den Ottolander GJ, ten Pas JG (1979) Insulin requirements in diabetic patients treated with sulphinpyrazone. J Int Med Res 7(4):328–331
Su WY, Li Y, Chen X et al (2021) Ginsenoside Rh1 Improves Type 2 diabetic nephropathy through AMPK/PI3K/Akt-mediated inflammation and apoptosis signaling pathway. Am J Chin Med 49(5):1215–1233
Butterfield RJ, Foley AR, Dastgir J et al (2013) Position of glycine substitutions in the triple helix of COL6A1, COL6A2, and COL6A3 is correlated with severity and mode of inheritance in collagen VI myopathies. Hum Mutat 34(11):1558–1567
Lau GJ, Godin N, Maachi H et al (2012) Bcl-2-modifying factor induces renal proximal tubular cell apoptosis in diabetic mice. Diabetes 61(2):474–484
Alvarez ML, DiStefano JK (2011) Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS ONE 6(4):e18671
Yan J, Yang X, Jiao X et al (2020) Integrative transcriptomic and proteomic analysis reveals CD9/ITGA4/PI3K-Akt axis mediates trabecular meshwork cell apoptosis in human glaucoma. J Cell Mol Med 24(1):814–829
Karamizadeh Z, Kamali Sarvestani E, Saki F et al (2013) Investigation of osteopontin levels and genomic variation of osteopontin and its receptors in Type 1 diabetes mellitus. J Endocrinol Invest 36(11):1090–1093
Ruddock MW, Stein A, Landaker E et al (2008) Saturated fatty acids inhibit hepatic insulin action by modulating insulin receptor expression and post-receptor signalling. J Biochem 144(5):599–607
Hu X, Wang S, Xu J et al (2014) Triterpenoid saponins from Stauntonia chinensis ameliorate insulin resistance via the AMP-activated protein kinase and IR/IRS-1/PI3K/Akt pathways in insulin-resistant HepG2 cells. Int J Mol Sci 15(6):10446–10458
Vlassara H, Striker GE (2013) Advanced glycation endproducts in diabetes and diabetic complications. Endocrinol Metab Clin North Am 42(4):697–719
Manigrasso MB, Juranek J, Ramasamy R et al (2014) Unlocking the biology of RAGE in diabetic microvascular complications. Trends Endocrinol Metab 25(1):15–22
Wada J, Makino H (2016) Innate immunity in diabetes and diabetic nephropathy. Nat Rev Nephrol 12(1):13–26
Chow F, Ozols E, Nikolic-Paterson DJ et al (2004) Macrophages in mouse type 2 diabetic nephropathy: correlation with diabetic state and progressive renal injury. Kidney Int 65(1):116–128
Vasavada N, Agarwal R (2005) Role of oxidative stress in diabetic nephropathy. Adv Chronic Kidney Dis 12(2):146–154
Dihingia A, Ozah D, Ghosh S et al (2018) Vitamin K1 inversely correlates with glycemia and insulin resistance in patients with type 2 diabetes (T2D) and positively regulates SIRT1/AMPK pathway of glucose metabolism in liver of T2D mice and hepatocytes cultured in high glucose. J Nutr Biochem 52:103–114
Funding
No funding support for this work at this time.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Ethical standard
This research does not involve ethical needs.
Informed consent
No informed consent was necessary for the study.
Additional information
This paper belongs to the Topical Collection "Diabetic Nephropathy", managed by Giuseppe Pugliese.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gui, H., Chen, X., Ye, L. et al. Seven basement membrane-specific expressed genes are considered potential biomarkers for the diagnosis and treatment of diabetic nephropathy. Acta Diabetol 60, 493–505 (2023). https://doi.org/10.1007/s00592-022-02027-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00592-022-02027-2