Abstract
Purpose
Aberrant expression of miRNAs has been demonstrated to contribute to human carcinogenesis. This study was aimed at profiling differentially expressed miRNAs in formalin-fixed and paraffin-embedded tissues of spinal chordoma and testing the potential for using altered expression of miRNAs as prognostic markers for spinal chordoma patients.
Methods
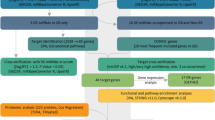
A miRNA array was used to profile differentially expressed miRNAs in spinal chordoma and nucleus pulposus tissues. Four of these differentially expressed miRNAs was then validated in spinal chordoma and control patients using quantitative RT-PCR. Bioinformatical analysis identified potential GO terms and signaling pathways affected by these microRNAs. Altered miR-1237-3p expression was then found to be associated with clinicopathological characteristics and prognosis of spinal chordoma patients.
Results
The miRNA arrays identified 29 differentially expressed miRNAs in spinal chordoma tissues, four of which were verified by qRT-PCR in 42 spinal chordomas and 14 control tissues. Bioinformatical analysis revealed that the potential target genes of these miRNAs were mainly involved in gene transcription, cell junction proteins, and gene pathways in cancer and endocytosis. Reduced miR-1237-3p expression was associated with tumor invasion and worse recurrence-free survival of spinal chordoma patients (χ 2 = 16.217, p = 0.000, log-rank test). Multivariate analyses showed that miR-1237-3p expression was an independent prognostic factor for patients with spinal chordoma (HR = 0.001, 95 % CI 0.000–0.136, p = 0.005).
Conclusion
The data from the current study identified a total of 29 differentially expressed miRNAs in chordoma tissues and reduced miR-1237-3p expression was associated with chordoma invasion and worse recurrence-free survival of the patients.



Similar content being viewed by others
References
Bydon M, Papadimitriou K, Witham T, Wolinsky JP, Bydon A, Sciubba D, Gokaslan Z (2012) Novel therapeutic targets in chordoma. Expert Opin Ther Targets 16(11):1139–1143
McMaster ML, Goldstein AM, Bromley CM, Ishibe N, Parry DM (2001) Chordoma: incidence and survival patterns in the United States, 1973–1995. Cancer Causes Control 12:1–11
Park L, Delaney TF, Liebsch NJ, Hornicek FJ, Goldberg S, Mankin H, Rosenberg AE, Rosenthal DI, Suit HD (2006) Sacral chordomas: impact of high-dose proton/photon-beam radiation therapy combined with or without surgery for primary versus recurrent tumor. Int J Radiat Oncol Biol Phys 65(5):1514–1521
Hsieh PC, Xu R, Sciubba DM, McGirt MJ, Nelson C, Witham TF, Wolinksy JP, Gokaslan ZL (2009) Long-term clinical outcomes following en bloc resections for sacral chordomas and chondrosarcomas: a series of twenty consecutive patients. Spine (Phila Pa 1976) 34(20):2233–2239
Fuchs B, Dickey ID, Yaszemski MJ, Inwards CY, Sim FH (2005) Operative management of sacral chordoma. J Bone Joint Surg Am 87(10):2211–2216
Akhavan-Sigari R, Gaab MR, Rohde V, Abili M, Ostertag H (2014) Expression of PDGFR-α, EGFR and c-MET in spinal chordoma: a series of 52 patients. Anticancer Res 34(2):623–630
de Castro CV, Guimaraes G, Aguiar S Jr, Lopes A, Baiocchi G, da Cunha IW, Campos AH, Soares FA, Begnami MD (2013) Tyrosine kinase receptor expression in chordomas: phosphorylated AKT correlates inversely with outcome. Hum Pathol 44(9):1747–1755
Hu H, Yang HL, Lu J, Chen KW, Qiu YH, Liu W, Luo ZP (2012) Association of telomerase expression with recurrence of sacral chordoma. Ann Oncol 23(10):2772
Horbinski C, Oakley GJ, Cieply K, Mantha GS, Nikiforova MN, Dacic S, Seethala RR (2010) The prognostic value of Ki-67, p53, epidermal growth factor receptor, 1p36, 9p21, 10q23, and 17p13 in skull base chordomas. Arch Pathol Lab Med 134(8):1170–1176
Shruti K, Shrey K, Vibha R (2011) Micro RNAs: tiny sequences with enormous potential. Biochem Biophys Res Commun 407(3):445–449
Inui M, Martello G, Piccolo S (2010) MicroRNA control of signal transduction. Nat Rev Mol Cell Biol 11(4):252–263
Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, Su TJ, Chiang CC, Li HN, Hong QS, Su HY, Chen CC, Chen WJ, Liu CC, Chan WK, Chen WJ, Li KC, Chen JJ, Yang PC (2008) MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell 13:48–57
Ozen M, Creighton CJ, Ozdemir M, Ittmann M (2008) Wide-spread deregulation of microRNA expression in human prostate cancer. Oncogene 27:1788–1793
Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ (2009) Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 58:1375–1381
Duan Z, Choy E, Nielsen GP, Rosenberg A, Iafrate J, Yang C, Schwab J, Mankin H, Xavier R, Hornicek FJ (2010) Differential expression of microRNA (miRNA) in chordoma reveals a role for miRNA-1 in Met expression. J Orthop Res 28(6):746–752
Bayrak OF, Gulluoglu S, Aydemir E, Ture U, Acar H, Atalay B, Demir Z, Sevli S, Creighton CJ, Ittmann M, Sahin F, Ozen M (2013) MicroRNA expression profiling reveals the potential function of microRNA-31 in chordomas. J Neurooncol 115(2):143–151
Long C, Jiang L, Wei F, Ma C, Zhou H, Yang S, Liu X, Liu Z (2013) Integrated miRNA-mRNA analysis revealing the potential roles of miRNAs in chordomas. PLoS One 8(6):e66676
Duan Z, Shen J, Yang X, Yang P, Osaka E, Choy E, Cote G, Harmon D, Zhang Y, Nielsen GP, Spentzos D, Mankin H, Hornicek F (2014) Prognostic significance of miRNA-1 (miR-1) expression in patients with chordoma. J Orthop Res 32(5):695–701
Zhang Y, Schiff D, Park D, Abounader R (2014) MicroRNA-608 and MicroRNA-34a regulate chordoma malignancy by targeting EGFR, Bcl-xL and MET. PLoS One 9(3):e91546
Chen K, Mo J, Zhou M, Wang G, Wu G, Chen H, Zhang K, Yang H (2014) Expression of PTEN and mTOR in sacral chordoma and association with poor prognosis. Med Oncol 31(4):886
Samson IR, Springfield DS, Suit HD, Mankin HJ (1993) Operative treatment of sacrococcygeal chordoma. A review of twenty-one cases. J Bone Joint Surg Am 75(10):1476–1484
Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ (2007) Cancer statistics, 2007. CA Cancer J Clin 57(1):43–66
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25(1):25–29
Shalgi R, Lieber D, Oren M, Pilpel Y (2007) Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput Biol 3(7):e131
Naka T, Kuester D, Boltze C, Schulz TO, Samii A, Herold C, Ostertag H, Roessner A (2008) Expression of matrix metalloproteinases-1, -2, and -9; tissue inhibitors of matrix metalloproteinases-1 and -2; cathepsin B; urokinase plasminogen activator; and plasminogen activator inhibitor, type I in skull base chordoma. Hum Pathol 39(2):217–223
Naka T, Boltze C, Kuester D, Schulz TO, Samii A, Herold C, Ostertag H, Roessner A (2004) Expression of matrix metalloproteinase (MMP)-1, MMP-2, MMP-9, cathepsin B, and urokinase plasminogen activator in non-skull base chordoma. Am J Clin Pathol 122(6):926–930
Xi Y, Nakajima G, Gavin E, Morris CG, Kudo K, Hayashi K, Ju J (2007) Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA 13(10):1668–1674
Deng ZQ, Yin JY, Tang Q, Liu FQ, Qian J, Lin J, Shao R, Zhang M, He L (2014) Over-expression of miR-98 in FFPE tissues might serve as a valuable source for biomarker discovery in breast cancer patients. Int J Clin Exp Pathol 7(3):1166–1171
Peiró-Chova L, Peña-Chilet M, López-Guerrero JA, García-Giménez JL, Alonso-Yuste E, Burgues O, Lluch A, Ferrer-Lozano J, Ribas G (2013) High stability of microRNAs in tissue samples of compromised quality. Virchows Arch 463(6):765–774
Li X, Lu Y, Chen Y, Lu W, Xie X (2013) MicroRNA profile of paclitaxel-resistant serous ovarian carcinoma based on formalin-fixed paraffin-embedded samples. BMC Cancer 13:216–223
Fu SW, Chen L, Man YG (2011) miRNA biomarkers in breast cancer detection and management. J Cancer 2:116–122
Zhang X, Chen J, Radcliffe T, Lebrun DP, Tron VA, Feilotter H (2008) An array-based analysis of microRNA expression comparing matched frozen and formalin-fixed paraffin-embedded human tissue samples. J Mol Diagn 10(6):513–519
Choi KS, Cohn MJ, Harfe BD (2008) Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: implications for disk degeneration and chordoma formation. Dev Dyn 237:3953–3958
Barry JJ, Jian BJ, Sughrue ME, Kane AJ, Mills SA, Tihan T, Parsa AT (2011) The next step: innovative molecular targeted therapies for treatment of intracranial chordoma patients. Neurosurgery 68(1):231–240 discussion 240-1
Diaz RJ, Cusimano MD (2011) The biological basis for modern treatment of chordoma. J Neurooncol 104(2):411–422
Di Fiore PP (2009) Endocytosis, signaling and cancer, much more than meets the eye. Preface. Mol Oncol 3(4):273–279
Polo S, Pece S, Di Fiore PP (2004) Endocytosis and cancer. Curr Opin Cell Biol 16(2):156–161
Lanzetti L, Di Fiore PP (2008) Endocytosis and cancer: an ‘insider’ network with dangerous liaisons. Traffic 9(12):2011–2021
Mellman I, Yarden Y (2013) Endocytosis and cancer. Cold Spring Harb Perspect Biol 5(12):a016949
Raitoharju E, Seppälä I, Oksala N, Lyytikäinen LP, Raitakari O, Viikari J, Ala-Korpela M, Soininen P, Kangas AJ, Waldenberger M, Klopp N, Illig T, Leiviskä J, Loo BM, Hutri-Kähönen N, Kähönen M, Laaksonen R, Lehtimäki T (2014) Blood microRNA profile associates with the levels of serum lipids and metabolites associated with glucose metabolism and insulin resistance and pinpoints pathways underlying metabolic syndrome: the cardiovascular risk in Young Finns Study. Mol Cell Endocrinol 391(1–2):41–49
Acknowledgments
We would like to thank OE Bio-tech (Shanghai, China) for technical support of this study. We also thank Medjaden Bioscience Limited (Hong Kong, China) for assistance in preparation of this manuscript.
Conflict of interest
The authors declare that there is no conflict of interest in this work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
586_2015_3927_MOESM2_ESM.jpg
Supplementary material 2. Analysis of miRNA–gene GO network. The blue box represents down-regulated miRNA-repressed gene (mRNA), red square represents down-regulated miRNA, and green square represents significant GO terms regulated by these miRNAs. The relationship between miRNAs, target genes and GOs are represented by gray lines. (JPEG 1333 kb)
586_2015_3927_MOESM3_ESM.jpg
Supplementary material 3. Analysis of miRNA–gene network. The blue box represents up-regulated miRNA-repressed genes (mRNA) and the red square represents up-regulated miRNA. The relationship between miRNAs and the target genes are represented by gray lines. (JPEG 3051 kb)
Rights and permissions
About this article
Cite this article
Zou, Mx., Huang, W., Wang, Xb. et al. Reduced expression of miRNA-1237-3p associated with poor survival of spinal chordoma patients. Eur Spine J 24, 1738–1746 (2015). https://doi.org/10.1007/s00586-015-3927-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00586-015-3927-9