Abstract
Since 2020, novel coronavirus pneumonia has been spreading rapidly around the world, bringing tremendous pressure on medical diagnosis and treatment for hospitals. Medical imaging methods, such as computed tomography (CT), play a crucial role in diagnosing and treating COVID-19. A large number of CT images (with large volume) are produced during the CT-based medical diagnosis. In such a situation, the diagnostic judgement by human eyes on the thousands of CT images is inefficient and time-consuming. Recently, in order to improve diagnostic efficiency, the machine learning technology is being widely used in computer-aided diagnosis and treatment systems (i.e., CT Imaging) to help doctors perform accurate analysis and provide them with effective diagnostic decision support. In this paper, we comprehensively review these frequently used machine learning methods applied in the CT Imaging Diagnosis for the COVID-19, discuss the machine learning-based applications from the various kinds of aspects including the image acquisition and pre-processing, image segmentation, quantitative analysis and diagnosis, and disease follow-up and prognosis. Moreover, we also discuss the limitations of the up-to-date machine learning technology in the context of CT imaging computer-aided diagnosis.
Similar content being viewed by others
1 Introduction
In December 2019, the first case of novel coronavirus pneumonia (COVID-19) was confirmed in Wuhan, China, which, nowadays, has spread rapidly worldwide and caused a global outbreak due to its strong infectiousness and high transmission rate [1]. The typical clinical symptoms of COVID-19 include fever, cough and malaise, with a few patients going upper respiratory and gastrointestinal symptoms such as nasal congestion, runny nose and diarrhea. Severe cases may develop complications such as acute respiratory distress syndrome, multi-organ failure, and even life-threatening [2,3,4]. Virus detection is critical for cutting off the transmission route, and the detection method includes the reverse transcription polymerase chain reaction (RT-PCR), i.e., nucleic acid testing. However, the limitations of nucleic acid detection exist, such as accuracy differences among manufacturers, poor sensitivity for patients with low viral load and difficulty in proper clinical sampling [5]. These issues make nucleic acid testing time-consuming and possibly false-negatives, which can easily lead to missed diagnoses and repeated testing [6, 7]. With the development of new medical imaging devices and technologies, many tools have been applied for disease diagnosis and evaluation, including X-ray, computed tomography (CT), cone-beam CT, magnetic resonance imaging, functional magnetic resonance imaging (MRI), ultrasound imaging, and so on. Among them, X-ray and CT are the two most commonly used diagnostic methods for COVID-19 analysis. X-ray is convenient and fast, and the examination cost is low. It can preliminatively determine whether a lesion occurs in a part of the patient’s body, but it is not easy to detect the specific details of early lesions, and the accuracy is not high. In contrast, CT is more sensitive and provides a clearer picture of microscopic lung lesions that cannot be detected by X-ray. It can show substantial lesions in COVID-19 patients, including ground glass shadow areas (GGO), interlobular septal thickening, prominent interlobular linens, and areas of consolidation [8,9,10]. It can help detect suspected cases earlier and is a highly effective method for COVID-19 detection. However, CT detection is prone to cross-infection, which requires good protection [7, 11]. In Italy, the USA and China, most cases of COVID-19 are identified by the presenting features in CT images [12]. However, the diagnostic accuracy of CT, with more than 300 images generated in a single scan and a high reading workload, depends to some extent on the experience and meticulousness of the physician. During the outbreak of COVID-19, thousands of CT images are produced while performing CT detection, and the prolonged manual review of the films is prone to misdiagnosis. Recently, the applications of machine learning technology in the field of medical imaging are increasing rapidly, which has dramatically eased the workload of clinical physicians while improving detection rate and diagnosis efficiency [13,14,15]. Especially, it has shown good results in using CT images to assist in lung disease analysis [16,17,18,19,20]. Machine learning techniques are widely applied to problems such as classification, detection, and segmentation in medical images. Computer-aided CT imaging diagnosis and treatment systems are of good detection sensitivity and automatic quantitative analysis, e.g., InferRead CT Pneumonia System (http://www.infervision.com/), Deepwise healthcare (www.deepwise.com), etc.
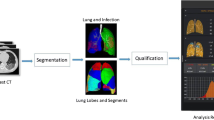
Generally, the procedure of computer-aided CT imaging diagnosis consists of (1) image acquisition and preprocessing, (2) image segmentation, (3) quantitative analysis and diagnosis, and (4) disease follow-up and prognosis. A large number of works have been studied for each process, such as Wang et al. [21] proposed an AI-assisted diagnosis and treatment system that automatically analyzes CT images and provides infection probabilities for rapid detection of COVID-19. It includes data collection, medical image segmentation and diagnostic functions that can save physicians about 30–40% of detection time. Shi et al. [22] proposed a machine learning-based method to distinguish COVID-19 from community-acquired pneumonia, which mainly includes steps such as image acquisition, pre-processing of lung infection and visual field, and calculation of infection area size distribution. Savitha et al. [23] proposed a fully automated computer-aided detection system with the functions of image noise reduction segmentation, feature extraction and selection, and lesion diagnostic classification. It can identify and differentiate subsolid nodules in the lungs shown in CT scans. Zhang et al. [24] developed an AI system to diagnose patients through lung lesion segmentation, accurate diagnosis of pneumonia, evaluation of drug treatment effects, and prognosis of critical diseases. Usually, the computer-aided diagnosis system acquires CT images of the patient’s chest. By importing these CT images, the system automatically segments the right and left lungs, lobes, and lung segments to identify inflammatory features and segment lesions in the lungs. The volume of the segmented lesions in the whole lung, the left and the right lung, and the corresponding percentage of infection are counted to determine whether the patient is infected with COVID-19.
2 ML-based CT imaging diagnosis
2.1 Image acquisition and preprocessing
The large volume of high-quality datasets, as the essentials for the model training, are urgently needed for the auxiliary diagnosis of COVID-19. Although some datasets are already available to public, the time-consuming task of labeling data, which usually takes 1–5 h for users to label out the infected regions of CT images, makes these existing public datasets very scarce compared to the data required for training. The common datasets currently available are shown in Table 1.
To improve the efficiency of data annotation, Shan et al. [25] proposed an artificial in-the-loop strategy and VB-Net semantic segmentation network model. The manual-in-the-loop strategy groups the training data. Meanwhile, the physician manually labels the smallest batch of CT data and uses the batch to train the segmentation network as an initial model to apply to the next dataset of infected regions. Then, the physician manually corrects the segmentation results and uses the corrected segmentation results as new training data input to iteratively increase the training dataset and builds the final VB-Net. Zhang et al. [25] proposed a deep domain adaptation method for diagnosing COVID-19 by transferring domain knowledge from the well-labeled common pneumonia source domain to the partially labeled COVID-19 target domain, minimizing domain differences by aligning the feature distributions of the two domains through adversarial learning and achieving 98.2% accuracy and 88.33% sensitivity using the ResNetl8 model. Yan et al. [26] mined these abundant retrospective medical data to build a large-scale lesion image dataset -DeepLesion, a dataset with 32,735 lesions in 32,120 CT slices from 10,594 studies of 4427 unique patients. Zheng et al. [27] proposed a deep learning algorithm to obtain strong detection performance without labeling training for COVID-19 lesions. The algorithm trains an unsupervised learning network to segment the lung region and then determines whether the infection with COVID-19 is present. The algorithm has great potential for clinical application in the accurate and rapid diagnosis of COVID-19, which is of important help to medical staff.
Although CT images are featured with higher image resolution compared with other imaging methods, images still need to be preprocessed before the segmentation. Messay et al. [28] used down-sampling algorithm to provide approximate resolution compatibility between test and training data before the segmentation of lung CT images. It significantly improved the efficiency of image processing and reduced the image noise. Local contrast enhancement was performed on each down-sampled profile image of a given case to improve image details. In order to avoid the constant power additive noise (e.g., Gaussian noise) in CT images hinders doctors’ diagnosis, Darmanayagam et al. [29] used Adaptive Wiener filter for denoising. Considering that there are various types of noises in the CT image processing, Chen et al. [30] used a median nonlinear filter with a 3 × 3 window to eliminate the noises, which was easy-to-obtain and was capable of keeping the image edge clear. In order to remove the effect of contrast difference caused by glare, noise and poor illumination conditions in the process of image acquisition. Ashwin et al. [31] generated a low-frequency image by replacing the pixel value with the median pixel value calculated on the 5 × 5 pixel square area centered on the pixel position and enhanced the CT image with the technology of limited contrast adaptive histogram equalization (CLAHE). Al-Tarawneh et al. [32] proposed a lung cancer detection technology, in which Gabor filtering, automatic enhancement and fast Fourier transform algorithm are used to preprocess lung CT images to achieve the purpose of eliminating noise, pollution and interference, respectively.
2.2 Image segmentation
Image pre-processing is an essential step for the computer-aided CT Imaging diagnosis and treatment of COVID-19. The presence of a large amount of non-essential information in the initial CT images can affect the efficiency and diagnostic accuracy of the system to some extent. Therefore, removing interference, such as chest tissue and image artifacts, and enhancing useful information are the main goal in this step. Segmenting the region of interest (ROI) in CT images through image processing can reduce the computation of subsequent algorithms while segmenting the lesion region can help track and observe the condition. The infection rate can be calculated based on the size of the infected region, which could assist in the follow-up of diagnosis and treatment. Image segmentation is a hot topic in the field of computer vision. Traditional image segmentation algorithms include threshold segmentation algorithm [33], edge detection segmentation algorithm [34], region segmentation algorithm [35], active contour model segmentation method [36], and graph cutting method [37], etc. These methods usually suffer from some issues, such as low computing speed, high noise level, and strongly influenced by subjective factors. In order to improve the accuracy and applicability of image segmentation, a large number of medical image segmentation methods based on machine learning are proposed (Fig. 1).
In 2012, Alex Krizhevsky first proposed AlexNet, the embryonic form of CNN [38], which completely adopted the training mode of supervised learning and adopted Relu activation function to accelerate the convergence rate. Dropout strategy is introduced to alleviate the over-fitting phenomenon in the training process. In 2014, Goodfellow proposed the generative adversarial network (GAN) [39] to complete the generation task under the condition of balance between generator and discriminator with the strategy of game training. In 2015, Evan Shelhamer et al. [40] replaced the full connection layer with the convolutional layer in CNN and proposed the full convolutional network (FCN). In 2015, Ronneberger et al. [41] designed u-Net network on the basis of FCN, which retained a large number of feature channels in the up-sampling process, so that more information could flow into the segmented image eventually restored. In order to facilitate the processing of 3D medical images and meet the needs of accurate segmentation of medical images, Milletari et al. [42] proposed a 3D deformation V-NET in 2016, which uses 3D convolution to check images for convolution operation. Zhou et al. [43] proposed u-Net++ network in 2018. Its advantage lies in the redesign of jump path and depth supervision, which can achieve rapid and accurate segmentation of lung region and promote the development of computer-aided diagnosis of lung diseases to a certain extent.
Chen et al. [44] proposed a U-Net ++ -based segmentation model for identifying COVID-19 on patients’ chest CT images. Firstly, the raw CT images were entered to the model, which processed them and outputted the predicted frames used to construct suspicious lesions. Then, valid regions were extracted and invalid fields were removed to avoid possible false positives. The CT images with the predicted results were divided into four quadrants. The results were output only when three consecutive images were predicted to have lesions in the same quadrant. The evaluated performance of the model achieved 95.2% accuracy, 100% sensitivity and 93.6% specificity. With the help of the model, radiologists’ reading time was reduced by 65%.
In 2015, Chen et al. [45] combined the idea of deep convolutional neural network (DCNN) and fully connected conditional random field (CRF) to construct the DeepLab model, which is capable of producing semantically accurate predictions and detailed segmentation graphs with computational efficiency. Based on the DeepLab, atrous convolution is used as a powerful tool in dense prediction tasks to reduce computational effort, it is a convolution idea proposed to reduce image resolution and lose information in image semantic segmentation. A spatial pyramid pool (ASPP) is proposed to segment objects at multiple scales, and finally, by combining DCNN and probabilistic graphical models, the localization of object boundaries is improved, and better semantic segmentation performance is achieved [46]. In 2017, Chen et al. revisited the application of atrous convolution in semantic image segmentation by designing cascaded or parallel atrous convolution modules to solve multiscale object segmentation problems and capturing multiscale contexts by employing multiple atrous rates. The previously proposed spatial pyramid pooling module is also extended to further improve the performance [47]. Based on the network of DeepLab V3, Chen et al. added a decoder network and proposed DeepLab V3+ . DeepLabv3+ adds a decoder model to DeepLabv3 and obtains an enhanced segmentation result, especially for object edges. It introduces the Xception in deep separable convolution and integrates the ASPP and decoder models to improve the training speed and robustness of encoder-decoder networks [48].
Alom et al. proposed the NABLA-N network in 2019 for the dermoscopic image segmentation task. This model ensures better feature representation for semantic segmentation by combining low-level to high-level feature maps, showing better quantitative and qualitative results with the same or fewer network parameters [49]. In 2020, Alom et al. applied the NABLA-N network to segment CT and X-ray images of patients with COVID-19, delineating the infected regions of the lungs [50].
In 2020, Guszt'av et al. [51] applied the U-Net architecture by adding Attention Gates (AGs) to improve the accuracy of the segmentation model, focusing on important local features and reducing the false positive rate by reducing the features with low correlation. A lung region segmentation network was designed using GAN and Attention U-Net to achieve 97.5% Dice on the JSRT dataset. Nabila et al. [52] proposed a generalized focal loss function based on the Tversky exponent to solve the problem of data imbalance in medical image segmentation. They improved the Attention U-Net model by adding an image pyramid to preserve contextual features and performed experiments on the BUS2017 dataset and ISIC2018 dataset, which improved the segmentation accuracy by 25.7% and 3.6%, respectively, compared with the raw U-Net.
Chen et al. [53] proposed a novel method for automatic segmentation of infected regions in CT images of patients with coronavirus pneumonia. The model is based on the U-Net structure, using the residual network to improve the segmentation of features, and constructing an efficient attention mechanism in the decoding process to achieve high-quality multi-class segmentation results and improve the effectiveness of U-Net. The experimental results show that the algorithm achieved an excellent performance in the automated segmentation of chest CT images for the COVID-19 patients.
Gozes et al. [54] developed an artificial intelligence-based automated CT image analysis tool for the detection, quantification and tracking of COVID-19. It was used to segment the lungs to extract ROI, a Resnet-50-2D deep convolutional neural network was used to detect novel coronavirus-related abnormalities, and commercial software was applied to analyze nodular or focal opacities in 3D space. Using the rapid evaluation of many CT images in different ways, a 95% accuracy rate was achieved in identifying COVID-19 in a public database in China. This primitive study is now being expanded to a larger population and could achieve high accuracy in COVID-19 detection as well as quantification and tracking of disease.
Rajinikanth et al. [55] propose an image-assisted system for extracting infected slices from lung CT images, which uses value filters to extract lung regions by eliminating possible artifacts, image enhancement by Harmony-Search-Optimization and Otsu thresholding. With the completion of segmentation to extract infected regions, the region of ROI is extracted from the binary image to assess the severity. Finally, the features extracted from the ROI are used to identify the pixel ratio between the lung and the infected part to determine the severity of the infection.
Kuchana et al. [56] proposed a two-dimensional deep learning framework with U-Net backbone for segmentation to identify interstitial lung spaces and modified the hyperparameters to improve the model’s performance and reduce the training time. Two segmentation tasks were performed using this framework: segmentation of lung gaps from CT slices; and segmentation of abnormal regions in chest CT scans associated with COVID-19, using semantic segmentation of lung CT images acquired by Kaggle and GitHub. Both segmentation tasks allow for chest CT scan prediction and volume assessment to provide statistical information on abnormalities associated with COVID-19.
Cao et al. [57] proposed a method based on the longitudinal progression of COVID-19 detection using deep learning at the voxel level to segment lung cloudiness. Different CT images from 10 positive cases of COVID-19 were used to predict manual segmentation. It uses a CNN-based U-Net with manual segmentation of lung images as a benchmark. The model was validated by analyzing the comparative evolution of two confirmed cases of COVID-19 in Wuhan.
Voulodimos et al. [58] proposed a deep learning-based CT image semantic segmentation method for identifying COVID-19 infected regions in patients’ CT images. The comparasive results of U-Net and FCN models for CT image segmentation showed that in the presence of data imbalance on the dataset and human labeling errors on the boundaries of symptom presentation regions, FCNN was still able to provide accurate segmentation of symptom regions, thus helping physicians to rapidly detect COVID-19 symptoms (Table 2).
2.3 ML-based COVID-19 diagnosis models
Computer-aided diagnostic tools are more mature in the diagnosis of pulmonary nodules than in the COVID-19. In 2016, Arnaud et al. [59] proposed a novel computer-aided detection system for pulmonary nodules using multi-viewpoint convolutional networks (ConvNets) from training data automatically learned discriminative features with a high detection sensitivity of 90.1% in 888 scans of the publicly available LIDC-IDRI dataset. In the same year, He et al. [15] proposed the residual network ResNet, which explicitly reformulates the layers as a learned residual function regarding the layer inputs. It is easier to optimize and can obtain accuracy from greatly increased depth, with only 3.57% error on the ImageNet test set. With this improvement, the depth of the neural network has been able to reach 152 layers. Jung et al. [60] achieved a competitive performance index score of up to 0.91 using a 3D deep convolutional neural network (3D DCNN) with shortcut connections and a 3D DCNN with dense connections to classify lung nodules in CT images. The system successfully mitigated the gradient disappearance problem using shortcut connectivity and dense connectivity by allowing fast and direct passage of gradients. Features of spherical nodules were captured more efficiently compared to shallow 3D CNNs (Fig. 2).
2.3.1 Methods for identifying the positive patient
Alom et al. [50] proposed a novel multi-task deep learning-based method for fast and efficient identification of COVID-19 patients based on the NABLA-N network segmentation model. It adopts the Inception Residual Recurrent Convolutional Neural Network and Transfer Learning (TL). The detection accuracy of this detection model is about 84.67% for X-ray images and 98.78% for CT images.
Mei et al. [61] combined chest CT presentation with clinical symptoms, exposure history and laboratory tests to rapidly diagnose positive COVID-19 patients. A deep convolutional neural network was developed to understand the imaging characteristics of patients with COVID-19. Then, support vector machine (SVM), random forest, and multilayer perceptron (MLP) classifier were used to classify the patients. Finally, a neural network model for COVID-19 is developed to predict patient status by combining predictive data and clinical information. The area under the curve AUC was 0.92 with a sensitivity of 84.3% when 279 patients were tested [61].
Wang et al. [21] compared widely used segmentation models such as full convolutional networks (FCN-8s), U-Net, V-Net, and 3D U-Net++, and advanced classification models (such as ResNet-50, Inception networks, DPN-92), and Attention ResNet-50. The “3D U-Net++ -ResNet-50” combination model was selected to achieve an optimal area under the curve AUC of 0.991, saving physicians approximately 30–40% of detection time.
Zheng et al. [62] proposed a 3D deep convolutional neural network (DeCoVNet) for detecting COVID-19 from CT images. The process includes three phases: The first phase is the network’s backbone, which consists of a normal 3D convolution, a batch specification layer and a pooling layer; the second phase consists of two 3D residual blocks (ResBlocks). In each block, a 3D feature map is passed to a 3D convolution with a batch specification layer and a shortcut connection containing the 3D convolution. The generated feature maps are added in an elemental way; the third stage is the progressive classifier (ProClf), consisting of three 3D convolutional layers and a fully connected layer (FC) containing the softmax activation function. ProClf progressively extracts information from CT images by 3D max-pooling and finally outputs the probability of positive and negative COVID-19 directly. However, in DeCoVNet, the network design and training process needs to be improved on 3D segmentation networks and using accurate ground facts annotated by experts. The generalizability of this approach should be validated by using databases from different hospitals rather than only one database.
Song et al. [63] constructed a new deep learning architecture DRENet. For a given CT image, a pre-trained ResNet50 was first combined with an FPN network to detect potential lesion regions at different scales. Based on the detected regions, the local features of each region and the relational features between regions are extracted again using ResNet50. These features are then connected with the global features extracted from the original images and fed into multilayer perception (MLP) for image-level prediction. Finally, the predictions of the patient’s CT images are aggregated and used for individual-level diagnosis. The model extracts the main lesion features, especially the gross glass opacity (GGO), to visually aid the physician in diagnosis.
Alshazly et al. [64] used a deep convolutional neural network architecture for a comprehensive study of COVID-19 detection based on CT images, exploring different CNN models to obtain the best performance. A migration learning technique is also proposed to provide the best performance using inputs tailored for each deep architecture. Using the t-SNE algorithm, the learning features were analyzed and the results showed well-separated clusters of non-COVID-19 and COVID-19 subjects. The obtained network was also analyzed using the Grad-CAM algorithm to obtain a high-resolution visualization showing abnormal differentiated regions in CT images, achieving 92.2% specificity, 93.7% sensitivity, 91.3% precision and 92.9% average accuracy.
Mukherjee et al. [65] designed a convolutional neural network CNN customized deep neural network DNN to test and train both chest X-ray and CT images. The system had a false negative rate value of 0.0208 and an AUC value of 0.9808, with an overall accuracy of 96.28%. In addition, the authors achieved better results when integrating chest X-ray and CT images to diagnose subjects with COVID-19 infection compared to conventional DNNs such as ResNet, MobileNet and InceptionV3.
Li et al. [66] proposed a stacked autoencoder detector model, which greatly improved the accuracy and recall of the detection model. Four autoencoders are first constructed as the first four layers of the whole stacked autoencoder detector model to extract better CT image features. Then, the four autoencoders are cascaded together and connected to the dense layer and softmax classifier to form the model. Finally, a new classification loss function is constructed by superimposing the reconstruction loss to improve the detection accuracy of the model. The model achieves an average accuracy of 94.7% and a recall of 94.1%.
Ardakani et al. [67] proposed a deep learning-based computer-aided diagnosis and treatment system using AlexNet, VGG-16, VGG-19, SqueezeNet, GoogleNet, MobileNet-V2, ResNet-18, ResNet-50 ResNet-101 and Xception that were used to distinguish whether patients were infected with COVID-19. The AUC of Xception was 0.994 (sensitivity, 98.04%; specificity, 100%; accuracy, 99.02%).
Han et al. [68] proposed an attention-based deep 3D multiple instance learning (AD3D-MIL). AD3D-MIL can semantically generate deep 3D instances after possible infected regions and further apply the attention-based pooling method to 3D instances to gain insight into the contribution of each instance to the label. It is shown that the algorithm achieves an overall accuracy of 97.9%, an AUC of 99.0%, and a Cohen kappa score of 95.7%, which can be an effective aid for COVID-19 screening.
Ko et al. [69] developed a fast-tracking COVID-19 classification network (FCONet) for diagnosing COVID-19 pneumonia based on a single chest CT image. FCONet was developed by migration learning using one of four state-of-the-art pre-trained deep learning models (VGG16, ResNet-50, Inception-v3, or Xception) as the backbone, and experimentally, ResNet-50 was shown to exhibit excellent diagnostic performance (99.58% sensitivity, 100.00% specificity, and 99.87% accuracy) and outperformed the other three pre-trained models in the test dataset.
Mishra et al. [70] experimentally evaluated various existing Deep CNN-based image classification methods VGG16, InceptionV3, ResNet50, DenseNet12 and DenseNet201 for identifying COVID-19 positive cases in chest CT images. A decision fusion-based approach combining the prediction of each individual Deep CNN model to improve the prediction performance is also proposed, and the experimental results show that the method exceeds 86% in all aspects of performance metrics while greatly reducing the number of false positives, which is highly practical in real-world diagnostic scenarios.
Wu et al. [71] constructed a multi-view fusion model based on a modification of the ResNet50 architecture, using a three-view image set instead of the RGB three-channel images in a typical ResNet50 as network input. The model achieved better performance through single-view model and subset analysis for initial screening of COVID-19 and showed great potential in improving diagnostic efficiency and reducing the workload of radiologists.
Jaiswal et al. [72] designed a novel deep migration learning model for COVID-19 with the help of a convolutional neural network and a pre-trained DenseNet201 model to diagnose whether a patient is infected or not. The proposed model, its learning weights on the ImageNet dataset, and convolutional neural structures were used to extract features. The performance of the proposed deep migratory learning model on chest CT images is evaluated through extensive experiments and compared with VGG16, ResNet152V2 and Inception-ResNetV2, the model classifies chest CT images with 99.82%, 96.25% and 97.4% accuracy for training, testing and validation, respectively, outperforming competing methods.
Yang et al. [73] developed a deep learning model based on the densely connected convolutional network (DenseNet) to detect COVID-19 features on patients’ high-resolution CT (HRCT) images. DenseNet is an improved CNN that can extract both shallow and internal features of images, showing excellent in ImageNet classification tasks performance. Experiments show that the model achieves a sensitivity of 97% and an accuracy of 92%, which is similar to the level of young radiologists, but with much higher detection efficiency.
2.3.2 Methods for distinguishing COVID-19 from other types of pneumonia cases
Lin et al. [74] developed a 3D deep learning framework COVID-19 for the robustness of detection. The COVNet framework consists of ResNet50 as the backbone network, using CT images as input and generating features for the corresponding images; features extracted from all images are merged using a maximum pooling operation; the final feature maps are sent to fully connected layers and softmax activation function to discriminate the probability scores of patient types. The model achieves a high sensitivity of 90% and a high specificity of 96% on an independent testing dataset, can accurately detect coronavirus 2019 and differentiate it from community-acquired pneumonia and other lung conditions.
Kang et al. [24] used a 3D classification network adapted from 3D ResNet-18 for patient diagnosis. The network uses multiple 3D convolutional blocks with residual connections to successively extract local and global contextual features and calculate the final prediction with a fully connected layer and a softmax activation function. COVID-19 was distinguished from other common pneumonia as well as normal controls with 92.49% accuracy, 94.93% sensitivity, 91.13% specificity and an area of 0.9797 under the subject’s working features on an internal validation dataset.
Cheng et al. [109] segmented lungs based on U-Net and identified positive cases of COVID-19 using a slice diagnostic module with 2D ResNet152 as the backbone. The parameters of ResNet152 were pre-trained on a considerable dataset, ImageNet. The output of the classification network four scores indicates the confidence level of four categories: non-pneumonia, community-acquired pneumonia, influenza A/B, and COVID-19. The sensitivity and specificity of the system for detecting COVID-19 were 0.8703 and 0.9660, respectively.
Xu et al. [75] proposed a method to automatically screen CT images of COVID-19 by deep learning techniques that can be used to differentiate between healthy cases, influenza A viral pneumonia IAVP, and COVID-19. A 3D deep learning model is first used to isolate candidate infection regions from the lung CT image set. These separated images are then classified into COVID-19, influenza A viral pneumonia IAVP and with confidence scores in the healthy group using a location-attention classification model. Finally, the type of infection and the overall confidence score were calculated for each CT case using the Noisy-OR Bayesian function, and the experimental results showed that the model was able to achieve an overall accuracy of 86.7% for detection.
Polsinelli et al. [76] proposed a lightweight convolutional neural network CNN design based on the SqueezeNet model for the differentiation of CT images of patients with COVID-19 from other community-acquired pneumonia or healthy CT images, which achieved 85.03% accuracy, 87.55% sensitivity, 81.95% specificity, 85.01% accuracy and 86.20% F1 score with accuracy comparable to more complex CNN designs, 10 times more efficient than more complex CNNs using pre-processing, and does not require GPU acceleration and is less demanding on equipment.
Shi et al. [22] proposed a machine learning method for extracting specific features of COVID-19 based on the infection size-aware random forest approach (iSARF), which can be used to distinguish COVID-19 from community-acquired pneumonia in patient CT images, resulting in a sensitivity of 90.7%, specificity of 83.3%, and accuracy of 87.9%. The system first uses a deep learning model to subdivide CT images into pulmonary and infected regions and then uses the generated predictive model to design a set of manually crafted location-specific features. Compared with models trained using CT-SS and radiomics features as well as other classifiers, the method proposed by Feng Shi et al. has excellent performance and is highly scalable for both thin-section and thick-section CT images. However, the system still suffers from insufficient data, a lack of real-time CT images and differential diagnoses of the severity and pneumonia subtypes.
Ozkaya et al. [77] proposed a new method for fusing and ranking deep features to detect early-stage COVID-19. The method obtained deep features by pre-training a convolutional neural network CNN model and VGG-16, GoogleNet and ResNet-50 were used as pre-training networks. To improve the performance, feature fusion and ranking methods were applied, and then, the processed data were classified using a support vector machine SVM. However, the lack of a large database and the failure to distinguish between common viral pneumonia and COVID-19 affected the diagnostic capability of the method to some extent.
Wang et al. [78] analyzed CT images based on an initial migration learning technique in which a GoogleNet Inception V3 CNN predefined model was used for training. The method was based on a retrospective multi-cohort diagnostic study using a modified model for differentiating COVID-19 from other common viral pneumonia. The method achieved high sensitivities of 0.88 and 0.83 on some internal and external CT image datasets, respectively, but still suffers from low signal-to-noise ratios and complex data integration, both of which pose challenges to its efficacy.
Liu et al. [79] developed an automated and robust deep learning model COVIDNet by directly analyzing 3D CT images that can rapidly and accurately distinguish COVID-19 from other pneumonia infections. COVIDNet is a modified DenseNet-264 model consisting of four dense blocks. Each dense block has a different number of combinatorial units. Each cell consists of two sequentially connected stacks with an instance normalization layer25, a ReLU activation layer, and a convolutional layer. Through dense connections, it receives feature maps from all previous cells in the same dense block. The training batch size is 8. An Adam optimizer26 with a learning rate of 0.001 is used to minimize the binary cross-entropy loss.
Bai et al. [80] built and evaluated an AI system for distinguishing COVID-19 from other common pneumonia by chest CT images, using the EfficientNet B4 architecture for the pneumonia classification task. They ultimately achieved 96% test accuracy, 95% sensitivity and 96% specificity, with results superior to those of general radiologists.
Ouyang et al. [81] developed a 3D CNN network with online attention improvement and a dual sampling strategy (i.e., “Attention RN34 +DS”) for differentiating community-acquired pneumonia from COVID-19 in chest CT images. The results show that the algorithm has an AUC value of 0.944, an accuracy of 87.5%, a sensitivity of 86.9%, a specificity of 90.1%, and an F1-score of 82.0%, which is suitable for assisting physicians in diagnosing COVID-19 in the early stages of the outbreak.
Wang et al. [82] proposed a framework to effectively predict whether CT images contain pneumonia while distinguishing COVID-19 from interstitial lung disease (ILD) caused by other viruses. Two 3D-ResNet-based branches are integrated into a model framework for end-to-end training by designing a prior attention residual learning (PARL) module to localize lesion regions better. Experimental results show that the proposed framework can significantly improve the performance of COVID-19 screening and be extended to other similar clinical applications, such as computer-aided detection and diagnosis of pulmonary nodules in CT images and glaucomatous lesions in retinal fundus images.
Yan et al. [83] designed an AI system using a multiscale convolutional neural network (MSCNN) and evaluated its performance at slice and scan levels. MSSP, MSCNN, and data augmentation were used together to alleviate the scarcity of training data to improve the diagnostic performance of the AI system. The experimental results show that the system has good diagnostic performance in detecting COVID-19 and distinguishes it from other common pneumonia with a limited amount of training data, which contributes significantly to help physicians make rapid diagnoses and reduce their heavy workload.
2.3.3 Methods for COVID-19 severity classification
Guan et al. [84] classified 244 patients into three categories: (1) mild; (2) common and (3) severe. They used semi-quantitative evaluation methods based on lobar and segmental CT scores, opacity-weighted scores, and quantitative evaluation methods for lesion volume to quantify lung lesions. All four quantification methods observed significantly higher lesion loads in the severe type than in the common type, and the semi-quantitative and quantitative methods had good reproducibility in the measurement of inflammatory lesions and were able to distinguish well between the common and severe patients. The quantitative method has high specificity and low sensitivity compared with the semi-quantitative method, which is highly sensitive and suitable for severe patients.
Tang et al. [85] proposed a random forest-based severity assessment model for COVID-19 to achieve an accurate severity assessment of COVID-19 using features from different perspectives and to explore potential features associated with the severity of COVID-19. The model inputs patients’ chest CT image data and radiomic features as well as laboratory indices of each patient to output the severity of COVID-19 in patients. It is found that some chest CT image features and laboratory indices are highly correlated with the severity of COVID-19 and are valuable for the clinical diagnosis of COVID-19. This is the first work to assess the severity of COVID-19 using chest CT images and laboratory indices.
Huang et al. [86] proposed the percentage pulmonary turbidity (QCT-PLO) parameter by using an automated deep learning approach to quantitatively assess lung load changes in patients with COVID-19 disease using continuous CT scans. The method was validated against data from 126 patients, and the CT lung cloudiness percentage was analyzed in 126 patients. It was observed that the quantitative lung parameters could be used to show the progression of COVID-19. However, their study still has limitations such as insufficient data, systematic confirmation that has not been proven to correlate directly with pathologic impact.
Shen et al. [87] proposed a quantitative CT analysis method for the severity stratification of COVID-19 suitable for the assessment of lesions, such as CT signs, consolidation of gross glass opacities (GGO), and significant fibrosis of the disease. They found that the proportion of solid lesions increased with the mean density of the lesion and the proportion of GGO decreased with the mean density of the lesion. Limitations of the method include the retrospective nature of the study, selection bias in cases of severe COVID-19, small sample size, and bias in assessment based on radiologist-defined CT scores. In the future, their work may be improved by examining the correlation between quantitative CT parameters and clinical symptoms, and laboratory clues will support clinical decision-making.
Chaganti et al. [88] proposed a method to automatically quantify gross glass shadows and solid lesion areas in chest CT scans using a deep learning algorithm. Using the patient’s chest CT as input, the lungs, lobes, and lesion areas were segmented three-dimensional. The percentage opacity (PO) and lung severity score (LSS) were calculated to quantify the extent of infected lungs and the distribution of infected lobes, respectively. However, they did not assess other lung diseases, including common pneumonia, making it difficult to distinguish other diseases from COVID-19 and the impact of this assessment process on the diagnostic power of the algorithm.
Gozes et al. [89] proposed a weakly supervised deep learning method based on chest images to detect, localize and quantify the severity of COVID-19 manifestations from patients’ chest CT images. The region of interest ROI of the lung is first localized in the patient’s chest CT. The second step classifies the lung ROI into normal and abnormal using a 2D ROI classification network. A fine-grained map of the pathological tissue is extracted using a multiscale application of the GradCam method. Finally, in order to understand the different patterns of abnormal disease manifestations, unsupervised clustering of normal and abnormal slices is proposed. However, the database of this system was formed only on the normal and abnormal regions of COVID-19, without classifying other viral types of pneumonia, so this may lead to a false positive diagnosis of COVID-19 cases.
Yu et al. [90] used four pre-trained deep models (Inception-V3, ResNet-50, ResNet-101, DenseNet-201) to extract features from CT images. They then fed these features to multiple classifiers (linear discriminant, linear SVM, cubic SVM, KNN, and Adaboost decision trees) to detect severe and non-severe COVID-19 cases. Three validation strategies (hold-validation, tenfold cross-validation, and leave-one-out method) were used to verify feasibility. The best performance was finally achieved by DenseNet-201 with a cubic SVM model (Table 3).
2.4 ML-based COVID-19 disease prognosis
In addition to the determination of COVID-19, disease prognosis are important in the management of COVID-19 treatment in order to monitor the progress of patients and investigate potential sequelae after treatment. The studies focused on disease prognosis are still limited.
2.4.1 Image-feature-oriented ML models
United Imaging Intelligence (UII) proposed machine learning-based methods and visualization techniques to verify changes in lesion size, density, and other clinically relevant factors in infected areas of patients with COVID-19 and then automatically generate diagnostic reports to reflect these changes and provide theoretical references for physicians to determine future treatment procedures. In addition, the Perception Vision Company (PVmed) team attempted to create a comparative model to reflect changes in different CT images of the same patient by aligning the infected areas and observing trends in these quantitative values, providing a follow-up solution for COVID-19 [91].
Qi et al. [92] developed and tested a machine learning-based CT radiomics model for predicting the length of stay in patients with COVID-19. Lesion regions in CT images were extracted using a U-net-based algorithm and fed into two machine learning models, logistic regression and random forest, which were trained and validated against each other by using a multicenter cohort on a separate set of data from the COVID-19 dataset. Their study has limitations such as small sample diversity and semi-automatic lesion segmentation that may lead to selection bias. A large prospective multicenter cohort is needed to properly set up and test the machine learning-based CT radiomics model.
Wang et al. [93] proposed a method for a fully automated deep learning system for diagnosis and prognostic analysis of COVID-19 using CT images. The model uses a DenseNet-like structure consisting of four dense blocks, where each dense block is a multiple stack of convolution, batch normalization, and ReLU activation layers. Dense connections are used inside each dense block to consider multiple layers of image information. A global averaging pool is used at the end of the last convolutional layer to generate 64-dimensional DL features. The final output neuron is fully connected to the DL features to predict the input patient’s probability of being infected with COVID-19. The 64-dimensional DL features are combined with clinical features such as age, gender, and concomitant disease to construct a combined feature vector, and prognostic features are selected using a stepwise approach. These selected features were subsequently used to construct a multivariate Cox proportional risk model [110] to predict the length of hospital stay required for patients to return to health.
2.4.2 Clinical-feature-oriented ML models
Jiang et al. [94] combined features such as lymphocyte count, white blood cell count, body temperature, creatinine, and hemoglobin to assess the probability of developing a fatal complication, acute respiratory distress syndrome ARDS, in patients with COVID-19. The system used machine learning model types such as decision trees, random forests, and support vector machines to achieve 80% accuracy.
Yan et al. [95] developed a novel prognostic model for coronavirus pneumonia based on the xGBoost (eXtrellle Gmdient Boosting) algorithm, which identified three key clinical features such as lactate dehydrogenase (LDH), lymphocytes and high-sensitivity C-reactive protein (hs-CRP) from more than 300 features for rapid prediction of patient risk, with an accuracy rate of over 90%. Enables detection, early intervention and potential reduction in mortality in high-risk patients. Enables high-risk patients to be prioritized for treatment and improves survival of patients with COVID-19.
Assaf et al. [96] used three different machine learning algorithms: neural networks, random forests, and classification and regression decision trees (CRT) to predict patient conditions changes. Significant differences were found between critically ill and non-critically ill patients with advanced COVID-19, mainly in vital signs (respiratory rate and room air oxygen saturation) and markers of inflammation (blood leukocytes, neutrophil count and CRP) and in APACHE II scores combining these markers. Machine learning algorithms amplify these markers’ diagnostic accuracy and discriminatory efficacy, maximizing their use to predict the risk of developing the severe disease in patients with COVID-19 over the course of treatment.
Zhang Li et al. [97] developed a fully automated AI system to quantify pulmonary abnormalities associated with patients with COVID-19 and to assess disease severity and disease progression using thick-layer chest CT images. This study found that a deep learning model trained on a multicenter community-acquired pneumonia CT dataset could be directly applied to segment lung abnormalities in patients with COVID-19; the fraction of infection (POI) and mean infectious HU (iHU) in severe and non-severe patients, with AUCs of 0.97 (95% CI 0.95, 0.98) and 0.69 (95% CI 0.63, 0.74), with a statistically significant difference (p value < 0.001); the change in POI evaluation of whole lung infections was in good agreement with the radiologists’ reports.
Liang et al. [98] combined deep learning techniques with traditional Cox models to perform a survival analysis of nonlinear effects of clinical covariates to predict clinical outcomes in patients with COVID-19. The C index and AUC of this method were 0.894 (0.95 CI, 0.857–0.930) and 0.911 (0.95 CI, 0.875–0.945), respectively, on the model validation set, demonstrating that this deep learning survival Cox model can efficiently classify patients with COVID-19 for high-precision classification.
Iwendi et al. [99] proposed a fine-tuned random forest model based on the AdaBoost algorithm. The model uses the travel history, health status, and demographic data of patients with COVID-19 to predict the severity of the patients and the subsequent possible outcomes. On the dataset used, the model had an accuracy of 94% and an F1 score of 0.86.
Abdulaal [100] et al. proposed an artificial neural network ANN that can analyze a range of patient characteristics, including demographics, comorbidities, lifestyle factors, and presenting symptoms, to predict the risk of death for patients during their current hospitalization. These data can be collected during the patient’s initial contact with the physician, thus allowing early prediction of outcomes in order to inform clinical management decisions as early as possible.
Cheng et al. [101] developed a new supervised machine learning classifier that can use hospital EMR data to predict the risk of transferring patients with COVID-19 to the ICU within the next 24 h. The system applies a random forest approach, is promising in analyzing multiple types of complex clinical data, has high model generalization, and can elucidate higher-order interactions between variables without compromising prediction accuracy, facilitating hospital planning and operational efficiency.
2.4.3 Clinical and image features mixed ML models
Zhang et al. [24] analyzed clinical data and radiological features leading to the progression of critical illness and developed AI-assisted models to estimate clinical prognosis. Systematically extracted quantitative pulmonary disease lesion characteristics and clinical parameters such as age, serum albumin, and oxygen saturation were created and applied to predict clinical outcomes from initial admission to progression to critical illness (by death or clinical need for mechanical ventilation or transfer to the ICU). The model used an optical gradient elevator and a Cox proportional risk regression model for prognosis prediction. The results showed that pulmonary lesions and clinical metadata could significantly predict prognosis. In conclusion, the work examining the prognosis of disease tracking in COVID-19 is still not very well established compared to the work on disease determination. However, with the continuous advancement of COVID-19 treatment tools and machine learning techniques, it is believed that more techniques and systems will be available for COVID-19 disease tracking and prognosis.
Ma et al. [102] used machine learning methods random forest and XGboost to analyze data on symptoms, complications, demographics, vital signs, CT scan results, and laboratory findings at the time of patient admission to predict clinical characteristics of the risk of death in patients with COVID-19.
Bai et al. [103] used a multilayer perceptron MLP and LSTM to build an artificial intelligence model to predict disease progression in mild patients with COVID-19 and found that the complementary nature of clinical data and quantitative chest CT sequences was important for predicting patient disease. The method can effectively and accurately identify mild patients prone to deteriorate into severe cases, which can help optimize treatment strategies, reduce mortality, and alleviate medical stress (Table 4).
3 Limitations
3.1 Data issue while applying ML to CT imaging diagnosis
Generally, the larger dataset a machine learning model uses for training, the better the performance of the system will achieve. However, although some datasets are publicly available, the sample number of effectively labeled COVID-19 CT images is still limited or they are of weak quality, resulting in a lack of qualified large-scale dataset for quantitative studies. Therefore, a database with a systematic review of submitted data is more conducive to computer-aided diagnosis and treatment system than the immediate release of data of weak quality as a public database [104]. Due to the high cost of sign sample image generation, the size of current sign libraries is around a hundred to a thousand images. The criteria for generating them vary among libraries and their coverage is limited. For training robust visual feature classification models, the samples are not sufficient, much less for the application of deep learning algorithms.
The NABLA-N network proposed by Alom et al. [50] was trained on only 300 samples in its initial version. The system output showed some false positives due to the lack of labeled samples for lung segmentation of CT images. Training and testing with more samples are needed to generalize and increase the accuracy of the model. The CT image semantic segmentation approach proposed by Voulodimos et al. [58] suffers from the data issues (such as the poor availability and the imbalance), affecting the complexity of the selection and topology of the classification model. Lin et al. [74] developed a 3D deep learning framework- COVNet, due to the lack of laboratory confirmation of the origin of pneumonia cases caused by other types of viruses, such as non-influenza viruses, bacterial pneumonia, and tissue pneumonia caused by any cause, and the fact that the testing and training datasets were from the same hospital, other viral pneumonia could not be selected for comparison in this study. Cheng et al. [109] proposed a 2D ResNet152 module that lacks more data on other subtypes of lung disease and the diagnostic capability of the system needs to be improved. The construction of a large dataset containing relevant CT and clinical information, especially for the underlying disease, would allow additional analysis of the diagnostic system and the development of more functionalities such as mortality severity assessment.
The small sample size is the main shortcoming of the deep convolutional neural network developed by Mei et al. [61]. Although the result of screening patients using this model is promising, further data collection is needed to test the model’s generalizability to other patient groups. The “3D U-Net++ -ResNet-50” combined model used by Wang et al. [21] relies too much on fully annotated CT images and a large set of annotated CT images, including lung contours, lesion regions and classification, is needed to train the network. The evaluation model proposed by Tang et al. [85] may lead to overfitting problem in the training phase of the model due to the few number of patients. The U-Net-based model- DeCoVNet for lung segmentation in the 3D deep convolutional neural network [62] did not utilize temporal information, was trained using an imperfect ground-truth mask and the data used in the study were from a single hospital without cross-validation. The number of model samples in the automatic screening method proposed by Xu et al. [75] was limited. The number of test samples should be expanded, and more multiple-center clinical studies should be conducted to improve the accuracy of the system, cope with complex clinical situations, and improve the segmentation and classification accuracy of the model. The GoogleNet Inception V3 CNN predefined model used by Wang et al. [78], with a low signal-to-noise ratio and complex data integration, poses a challenge to its efficacy. The performance of this system will increase when the training volume increases. To optimize the diagnostic system, a large database is needed to link it to all pathological stages of disease progression and COVID-19. DRENet proposed by Song et al. [63] could not effectively address batch effects or make accurate predictions for other data sources due to insufficient training data. The method works efficiently only on the original data of the hospital dataset but could not directly predict external data (i.e., not produced by the same hospital). The COVID-19 classification network FCONet developed by Ko et al. [69] was mainly validated using split-test datasets. Therefore, the test dataset obtained from the same source as the training dataset may trigger generalization and overfitting problems. Yan et al. [83] designed an AI system that worked properly on a test dataset of 88 CT scans but still needed to be tested on a large CT dataset to prove its generalization.
3.2 Clinic issues while applying ML to CT imaging diagnosis
Computer-aided diagnosis and treatment system faces many difficulties in clinical use. The structure of body organs is complex, e.g., the lung has many branches. Its internal structure shows polymorphism, and the grayscale of various tissues on medical images is similar. Therefore, the responses to various types of lung injuries can easily overlap. There is also a great deal of overlap in presenting many lung diseases that depend on factors, e.g., age, drug reactions, immune status, and potential complications. Clinical practice shows that the current pneumonia auxiliary diagnosis system cannot accurately recognize the main signs of pneumonia, such as grinded-glass shadow, solid shadow, striated shadow, and lobular septal thickening. Moreover, the clinical features of COVID-19 and other common pneumonia (e.g., community-acquired pneumonia, histoplasmosis, and eosinophilic pneumonia) are similar. The CT imaging manifestations have some overlap and low differentiation, which is easy to cause misdiagnosis and omission in the clinical diagnosis.
COVNet, a 3D deep learning framework developed by Lin et al. [74], is difficult to distinguish all lung diseases only based on the imaging presentation of chest CT scans. Given the non-specific nature of the grinded-glass opacity and other features on chest CT images, the AI model proposed by Mei et al. [61] has some limitations in its ability to discriminate COVID-19 from other causes of respiratory failure. Therefore, the algorithm may be helpful in places with a high prevalence of COVID-19, but may not work accurately in places or times with low prevalence. Wang et al. [21] used a combined “3D U-Net++ -ResNet-50” model that performed poorly in multiple types of lesions or with significant metallic or motion artifacts. Xu et al. [75] proposed an automated screening method for CT images of COVID-19 that could not distinguish COVID-19 from other common pneumonia. The accuracy of the 3D CNN network developed by Ouyang et al. [81] in identifying COVID-19 infections in small regions was not satisfactory. Therefore, the clinical diagnosis of COVID-19 still requires the combination of CT images and other information, such as the patient’s exposure history, travel history and onset symptoms. The system designed to automatically detect quantitative COVID-19 CT images has difficulty in distinguishing COVID-19-associated Abnormalities from other types of diseases such as interstitial lung disease, non-solid pulmonary nodules and heart failure [105]. Moreover, the presence of co-morbidities may affect the segmentation and assessment of CT imaging, and it usually requires additional procedures to complete the task. In addition, these methods cannot accurately detect tiny areas close to blood vessels or very low-density pulmonary grinded-glass shadows. In practice, clinical information or other tests should be considered along with imaging findings to rule out false-positive detection at the final diagnosis [107].
3.3 Interpretability while applying ML to CT imaging diagnosis
A common issue of all machine learning approaches is the lack of transparency and interpretability [106], which could potentially pose a severe threat and cause some constraints in practical applications. Computer-assisted diagnosis and treatment systems are usually used only as an auxiliary tool, and a qualified assisted diagnosis and treatment system must be transparent and interpretable to gain the trust of the physicians and patients. A model that lacks interpretability may lead to unreliable treatment solution for patients and, in some cases, threatens patients’ life.
COVNet developed by Lin et al. [74] uses heat maps to visualize important regions of the scan that lead to algorithmic decisions. However, the heat maps are still not sufficient to model which unique features will be used to differentiate COVID-19 from community-acquired pneumonia. DeCoVNet [62] works in a black-box fashion in diagnosing COVID-19, and its interpretability is still at an early stage. In the 3D CNN network developed by Ouyang et al. [81], although the proposed online attention module can greatly improve the interpretability of COVID-19 diagnosis, but (the same as traditional methods, e.g., Grad-CAM) it also needs to analyze the correlation between these attentional localizations and specific imaging signs commonly used in clinical diagnosis. A limitation of the MSCNN by Yan et al. [83] using multiscale convolutional neural networks comes from the black box: Although attention maps aid interpretation by highlighting significant regions, they are still insufficient to visualize the unique features of CNN algorithms used to distinguish COVID-19 from community-acquired pneumonia (Table 5).
4 Conclusion
Although the research of machine learning applied to medical diagnosis and treatment systems has achieved significant advancement, the computer-aided diagnosis and treatment systems based on CT imaging has not yet been widely adopted in hospitals. There are still many issues that need to be addressed. More high-quality datasets should be collected and applied to improve the algorithm performance. Machine learning methods such as weakly supervised, self-supervised, and deep migration can be used to train samples and alleviate the problem of insufficient data. In addition, clinical data, travel exposure history, can also be taken into the diagnosis to make up for the shortage of medical images. The interpretability has been an critical issue while applying AI in health care industry, and it ensures the impartiality of decision-making and promotes system stability by highlighting potential adversarial perturbations that may alter predictions, ensuring the presence of potentially true causal relationships in model inference. Recently, a number of researchers have proposed interpretable artificial intelligence (XAI), which is able to achieve finer localization mapping than traditional class activation mapping (CAM) while highlighting important regions that are closely related to predicted outcomes. It also can facilitate the use of AI-assisted diagnosis in clinical practice and contribute to the trust and transparency of AI systems, which are considered to be essential for AI to continue steadily develop without disruption [107, 108].
CT imaging technology played an important role in the diagnosis of COVID-19. This paper comprehensively discussed the applications of machine learning in aiding the diagnosis and treatment of COVID-19 and analyzed existing literature works published by global scholars in data acquisition, image processing, disease diagnosis, tracking prognosis, etc. The adoption of CT imaging computer-aided diagnosis and treatment systems is limited by the issues such as small sample size, the lack of detailed application scenarios and quantitative assessment, the unsatisfactory identification of the main signs of pneumonia, and the low discrimination. In future, there is still tremendous space for developing datasets, comprehensive diagnostic data, and interpretable artificial intelligence methods. Establishing a faster and more accurate computer-aided diagnosis and treatment system can better assist doctors in detecting, diagnosing, and treating COVID-19 and other diseases, improving the efficiency of diagnosis and treatment and the cure rate, and has a broad clinical application prospect.
References
Hui D et al (2020) The continuing COVID-19 epidemic threat of novel coronaviruses to global health-the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis 91:264–266
Huang C, Wang Y, Li X et al (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395(10223):497–506
Chen N, Zhou M, Dong X et al (2020) Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395(10223):507–513
Li Q, Guan X, Wu P et al (2020) Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 382(13):1199–1207
Fang Y, Zhang H, Xie J et al (2020) Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology 296(2):E115–E117
Ai T, Yang Z, Hou H et al (2020) Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology 296(2):E32–E40
Xie X, Zhong Z, Zhao W et al (2020) Chest CT for typical coronavirus disease 2019 (COVID-19) pneumonia: relationship to negative RT-PCR testing. Radiology 296(2):E41–E45
Ufuk F, Savaş R (2020) Chest CT features of the novel coronavirus disease (COVID-19). Turk J Med Sci 50(4):664–678
Zhang S, Li H, Huang S et al (2020) High-resolution computed tomography features of 17 cases of coronavirus disease2019 in Sichuan province, China. Eur Respir J 55:2000334
Wang L, Gao Y, Zhang G (2020) The clinical dynamics of 18 cases of COVID-19 outside of Wuhan, China. Eur Respir J 55:2000398
Pan Y, Guan H, Zhou S et al (2020) Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan, China. Eur Radiol 30(6):3306–3309
Kumar M, Gupta S, Kumar K, Sachdeva M (2020) Spreading of COVID-19 in India, Italy, Japan, Spain, UK, US: a prediction using ARIMA and LSTM model. Digit Gov: Res Pract 1(4):1–9
Suzuki K (2017) Overview of deep learning in medical imaging. Radiol Phys Technol 10(3):257–273
Coudray N, Ocampo P, Sakellaropoulos T et al (2018) Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 24(10):1559–1567
He K, Zhang X, Ren S & Sun J (2015) Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In: Proceedings of the IEEE international conference on computer vision, pp 1026–1034
Wang S, Shi J, Ye Z et al (2019) Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J 53:1800986
Walsh S, Calandriello L, Silva M et al (2018) Deep learning for classifying fibrotic lung disease on high-resolution computed tomography: a case-cohort study. Lancet Respir Med 6:837–845
Walsh S, Humphries S, Wells A, et al (2020) Imaging research in fibrotic lung disease; applying deep learning to unsolved problems. Lancet Respir Med (in press)
Angelini E, Dahan S, Shah A (2019) Unravelling machine learning: insights in respiratory medicine. Eur Respir J 54:1901216
Wang S, Zhou M, Liu Z et al (2017) Central focused convolutional neural networks: developing a data-driven model for lung nodule segmentation. Med Image Anal 40:172–183
Jin S, Wang B, Xu H, Luo C, Wei L, Zhao W et al (2020) AI-assisted CT imaging analysis for COVID-19 screening: Building and deploying a medical AI system in four weeks, MedRxiv
Shi F, Xia L, Shan F et al (2020) Large-scale screening of covid-19 from community acquired pneumonia using infection size-aware classification. arXiv:2003.09860
Savitha G, Jidesh P (2019) A fully-automated system for identification and classification of subsolid nodules in lung computed tomographic scans. Biomed Signal Process Control 53:101586
Zhang K, Liu X, Shen J et al (2020) Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of covid-19 pneumonia using computed tomography. Cell 181(6):1423–1433
Shan F, Gao Y Z, Wang J et al (2021) Lung infecIion quantification of COVID-19 in CT images with deep learning. Med Phys 633–1645
Yan K, Wang X, Lu L, Summers R (2018) DeepLesion: Automated mining of large-scale lesion annotations and universal lesion detection with deep learning. J Med Imaging 5:1. https://doi.org/10.1117/1.JMI.5.3.036501
Zheng C, Deng X, Fu Q, Zhou Q, Feng J, Ma H, et al (2020) Deep learning-based detection for COVID-19 from chest CT using weak label, MedRxiv
Messay T, Hardie R, Rogers S (2010) A new computationally efficient CAD system for pulmonary nodule detection in CT imagery. Med Image Anal 14(3):390–406
Darmanayagam S, Harichandran K, Cyril S et al (2013) A novel supervised approach for segmentation of lung parenchyma from chest CT for computer-aided diagnosis. J Digit Imaging 26:496–509
Chen Z, Sun X, Nie S (2007) An efficient method of automatic pulmonary parenchyma segmentation in CT images. In: 2007 29th annual international conference of the IEEE Engineering in Medicine and Biology Society, pp 5540–5542
Ashwin S, Ramesh J, Kumar S, Gunavathi K (2012) Efficient and reliable lung nodule detection using a neural network based computer aided diagnosis system. Int Conf Emerg Trends Electr Eng Energy Manag (ICETEEEM) 2012:135–142
Al-Tarawneh M (2012) Lung cancer detection using image processing techniques. Leonardo Electron J Pract Technol 11(21):147–158
Helen R, Kamaraj N, Selvi K, Raman V (2011) Segmentation of pulmonary parenchyma in CT lung images based on 2D Otsu optimized by PSO. Int Conf Emerg Trends Electr Comput Technol 2011:536–541
Saad M, Muda Z, Ashaari N, Hamid H (2014) Image segmentation for lung region in chest X-ray images using edge detection and morphology. In: 2014 IEEE international conference on control system, computing and engineering (ICCSCE 2014), pp 46–51
Zhang X, Li X, Feng Y (2015) A medical image segmentation algorithm based on bi-directional region growing. Optik 126(20):2398–2404
Zhang Y, Matuszewski B, Shark L, Moore C (2008) Medical image segmentation using new hybrid level-set method. Fifth international conference biomedical visualization: information visualization in medical and biomedical informatics 2008:71–76
Linguraru M, Pura J, Pamulapati V, Summers R (2012) Statistical 4D graphs for multi-organ abdominal segmentation from multiphase CT. Med Image Anal 16(4):904–914
Krizhevsky A, Sutskever I, Hinton G (2017) ImageNet classification with deep convolutional neural networks. Commun ACM 60(6):84–90
Goodfellow I, Pouget-Abadie J, Mirza M et al (2014) Generative adversarial networks. Adv Neural Iinform Process Syst 3:2672–2680
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. IEEE conference on computer vision and pattern recognition (CVPR) 2015:3431–3440
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention, pp 234–241
Milletari F, Navab N, Ahmadi S (2016) V-net: Fully convolutional neural networks for volumetric medical image segmentation. In: 2016 fourth international conference on 3D vision (3DV), pp 565–571
Zhou Z, Siddiquee M, Tajbakhsh N, Liang J (2018) UNet++: A nested U-net architecture for medical image segmentation. Deep Learn Med Image Anal Multimodal Learn Clin Decis Support 2018(11045):3–11
Chen J, Wu L, Zhang J et al (2019) Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study. Sci Rep 10:1–27
Chen L, Papandreou G, Kokkinos I, Murphy K, Yuille A (2015) Semantic image segmentation with deep convolutional nets and fully connected CRFs. In: Proceedings of international conferences on learning and representations, 2015
Chen L, Papandreou G, Kokkinos I et al (2018) DeepLab: Semantic image segmentation with deep convolutional nets, atrous convolute on, and fully connected CRFs. IEEE Trans Pattern Anal Mach Intell 40(4):834–848
Zhou X, Liang W, Li W, Yan K, Shimizu S, Wang K (2021) Hierarchical adversarial attacks against graph neural network based IoT network intrusion detection system. IEEE Internet Things J. https://doi.org/10.1109/JIOT.2021.3130434
Chen LC, Zhu Y, Papandreou G et al (2018) Encoder-decoder with atrous separable convolution for semantic image segmentation. In: European conferenceon computer vision, pp 833–851
Alom MZ, Aspiras TH, Taha TM, Asari VK (2019) Skin cancer segmentation and classification with NABLA-N and inception recurrent residual convolutional networks. ArXiv, abs/1904.11126
Liang W, Hu Y, Zhou X, Pan Y, Wang K (2021) Variational few-shot learning for microservice-oriented intrusion detection in distributed industrial IoT. IEEE Trans Industr Inf. https://doi.org/10.1109/TII.2021.3116085
Ga'al G, Maga B, Lukács A (2020) Attention U-net based adversarial architectures for chest X-ray lung segmentation information. ArXiv, abs/2003.10304
Abraham N, Khan NM (2019) A novel focal tversky loss function with improved attention U-net for lesion segmentation. In: 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019), pp 683–687.
Chen X, Yao L, Zhang Y (2020) Residual attention u-net for automated multi-class segmentation of covid-19 chest CT images, pp 1–7, arXiv:2004.05645
Gozes O, Frid-Adar M, Greenspan H, Browning PD, Zhang H, Ji W, Bernheim A, Siegel E (2020) coronavirus (covid-19) pandemic: initial results for automated detection & patient monitoring using deep learning CT image analysis, pp 1–23, arXiv:2003.05037
Zhou X, Xu X, Liang W, Zeng Z, Yan Z (2021) Deep-learning-enhanced multitarget detection for end-edge-cloud surveillance in smart IoT. IEEE Internet Things J 8(16):12588–12596. https://doi.org/10.1109/JIOT.2021.3077449
Kuchana M, Srivastava A, Das R, et al (2020) AI aiding in diagnosing, tracking recovery of COVID-19 using deep learning on Chest CT scans. Multim Tools Appl 1–15
Cao Y, Xu Z, Feng J, Jin C, Han X, Wu H, Shi H (2020) Longitudinal assessment of covid-19 using a deep learning-based quantitative CT pipeline: illustration of two cases. Radiol Cardiothorac Imag 2:1–2
Athanasios Voulodimos, Eftychios Protopapadakis, Iason Katsamenis, Anastasios Doulamis, Nikolaos Doulamis (2021) Deep learning models for COVID-19 infected area segmentation in CT images. In: The 14th PErvasive Technologies Related to Assistive Environments Conference (PETRA 2021). Association for Computing Machinery, New York, NY, USA, pp 404–411
Setio AAA et al (2016) Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans Med Imaging 35(5):1160–1169
Jung H, Kim B, Lee I, Lee J, Kang J (2018) Classification of lung nodules in CT scans using three-dimensional deep convolutional neural networks with a checkpoint ensemble method. BMC Med Imaging 18(1):48
Mei X, Lee HC, Diao KY et al (2020) Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat Med 26(8):1224–1228
Zheng C, Deng X, Fu Q et al. Deep learning-based detection for COVID-19 from chest CT using weak label. https://doi.org/10.1101/2020.03.12.20027185.
Song Y, Zheng S, Li L, Zhang X, Zhang X, Huang Z, et al (2020) Deep learning enables accurate diagnosis of novel coronavirus (covid-19) with ct images. medRxiv, pp 1–10
Alshazly H, Linse C, Barth E, Martinetz T (2020) Explainable COVID-19 detection using chest CT scans and deep learning. Sensors 21:445
Mukherjee H, Ghosh S, Dhar A et al (2020) Deep neural network to detect COVID-19: one architecture for both CT Scans and Chest X-rays. Appl Intell 30:1–13
Li D, Fu Z, Xu J (2020) Stacked-autoencoder-based model for COVID-19 diagnosis on CT images. Appl Intell 30:1–13
Ardakani A, Kanafi A, Acharya U et al (2020) Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput Biol Med 121:103795
Han Z et al (2020) Accurate screening of COVID-19 using attention-based deep 3D multiple instance learning. IEEE Trans Med Imaging 39(8):2584–2594
Ko H, Chung H, Kang WS et al (2020) COVID-19 pneumonia diagnosis using a simple 2d deep learning framework with a single chest CT image: model development and validation. J Med Internet Res 22:e19569. https://doi.org/10.2196/19569
Mishra AK, Das SK, Roy P, Bandyopadhyay S (2020) Identifying COVID19 from chest CT images: a deep convolutional neural networks based approach. J Healthc Eng 2020:8843664
Wu X, Hui H, Niu M, Li L, Wang L, He B et al (2020) Deep learning-based multi-view fusion model for screening 2019 novel coronavirus pneumonia: a multicentre study. Eur J Radiol 128:109041
Jaiswal A, Gianchandani N, Singh D, Kumar V, Kaur M (2020) Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. J Biomol Struct Dyn 39:1–8
Yang S, Jiang L, Cao Z, Wang L, Cao J, Feng R et al (2020) Deep learning for detecting corona virus disease 2019 (COVID-19) on high-resolution computed tomography: a pilot study. Ann Transl Med 8:450. https://doi.org/10.21037/atm.2020.03.132
Li L, Qin L, Xu Z et al (2020) Using artificial intelligence to detect COVID-19 and community-acquired pneumonia Based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology 296(2):E65–E71
Xu X, Jiang X, Ma C et al (2019) A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering 6:1122–1129
Polsinelli M, Cinque L, Placidi G (2020) A light CNN for detecting COVID-19 from CT scans of the chest. Pattern Recognit Lett 140:95–100
Zhou X, Li Y, Liang W (2021) CNN-RNN based intelligent recommendation for online medical pre-diagnosis support. IEEE/ACM Trans Comput Biol Bioinformatics 18(3):912–921. https://doi.org/10.1109/TCBB.2020.2994780
Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, Cai M, et al (2020) A deep learning algorithm using ct images to screen for corona virus disease (covid-19). medRxiv pp 1–28
Liu B, Liu P, Dai L et al (2021) Assisting scalable diagnosis automatically via CT images in the combat against COVID-19. Sci Rep 11:4145. https://doi.org/10.1038/s41598-021-83424-5
Bai H, Wang R, Xiong Z, Hsieh B, Chang K, Halsey K et al (2020) Artificial intelligence augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other origin at chest CT. Radiology 296:E156–E165
Ouyang X et al (2020) Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans Med Imaging 39(8):2595–2605
Wang J et al (2020) Prior-attention residual learning for more discriminative COVID-19 screening in CT images. IEEE Trans Med Imaging 39(8):2572–2583
Yan T, Wong P, Ren H, Wang H, Wang J, Li Y (2020) Automatic distinction between COVID-19 and common pneumonia using multi-scale convolutional neural network on chest CT scans. Chaos Solitons Fractals 140:110153
Guan X, Yao L, Tan Y et al (2021) Quantitative and semi-quantitative CT assessments of lung lesion burden in COVID-19 pneumonia. Sci Rep 11(1):5148
Tang Z, Zhao W, Xie X et al (2021) Severity assessment of COVID-19 using CT image features and laboratory indices. Phys Med Biol 66(3):035015
Huang L, Han R, Ai T et al (2020) Serial quantitative chest CT assessment of covid-19: deep-learning approach. Radiol: Cardiothorac Imag 2:1–30
Shen C, Yu N, Cai S et al (2019) Quantitative computed tomography analysis for stratifying the severity of Coronavirus disease. J Pharm Anal 10:123–129
Chaganti S, Grenier P, Balachandran A, Chabin G, Cohen S et al (2020) Quantification of tomographic patterns associated with covid-19 from chest ct. arXiv:2004.01279 pp 1–24
Zhou X, Xu X, Liang W, Zeng Z, Shimizu S, Yang L, Jin Q (2022) Intelligent small object detection based on digital twinning for smart manufacturing in industrial CPS. IEEE Trans Indust Inform 18(2):1377–1386
Yu Z, Li X, Sun H, Wang J, Zhao T, Chen H et al (2020) Rapid identification of COVID-19 severity in CT scans through classification of deep features. Biomed Eng Online 19:63. https://doi.org/10.1186/s12938-020-00807-x
Shi F et al (2021) Review of artificial intelligence techniques in imaging data acquisition, segmentation, and diagnosis for COVID-19. IEEE Rev Biomed Eng 14:4–15
Qi X, Jiang Z, Yu Q, Shao C, Zhang H, et al (2020) Machine learning-based ct radiomics model for predicting hospital stay in patients with pneumonia associated with sars-cov-2 infection: a multicenter study. https://doi.org/10.1101/2020.02.29.20029603
Wang S, Zha Y, Li W, Wu Q, Li X, Niu M, Wang M et al (2020) A fully automatic deep learning system for covid-19 diagnostic and prognostic analysis. Eur Respir J 56(2):2000775
Jiang X, Coffee M, Bari A et al (2020) Towards an artificial intelligence framework for data-driven prediction of coronavirus clinical severity. Comput Mater Continua 62:537–551
Zhou X, Liang W, Wang K, Yang L (2021) Deep correlation mining based on hierarchical hybrid networks for heterogeneous Big Data recommendations. IEEE Trans Comput Soc Syst 8(1):171–178
Assaf D, Gutman Y, Neuman Y, Segal G, Amit S, Gefen-Halevi S et al (2020) Utilization of machine-learning models to accurately predict the risk for critical COVID-19. Intern Emerg Med 15:1435–1443
Li Z, Zhong Z, Li Y, Zhang T, Gao L, Jin D et al (2020) From community-acquired pneumonia to COVID-19: a deep learning–based method for quantitative analysis of COVID-19 on thick-section CT scans. Eur Radiol 30:6828–6837
Liang W, Yao J, Chen A, Lv Q, Zanin M, Liu J et al (2020) Early triage of critically ill COVID-19 patients using deep learning. Nat Commun 11:3543
Iwendi C, Bashir A, Peshkar A, Sujatha R, Chatterjee J, Pasupuleti S et al (2020) COVID-19 patient health prediction using boosted random forest algorithm. Front Public Health 8:357
Abdulaal A, Patel A, Charani E, Denny S, Mughal N, Moore L (2020) Prognostic modeling of COVID-19 using artificial intelligence in the united kingdom: model development and validation. J Med Internet Res 22:e20259
Cheng F, Joshi H, Tandon P, Freeman R, Reich D, Mazumdar M et al (2020) Using machine learning to predict ICU transfer in hospitalized COVID-19 patients. J Clin Med 9:1668
Ma X, Ng M, Xu S, Xu Z, Qiu H, Liu Y et al (2020) Development and validation of prognosis model of mortality risk in patients with COVID-19. Epidemiol Infect 148:e168
B Xiang, F Cong, Z Yu, et al. Predicting COVID-19 malignant progression with AI techniques. https://doi.org/10.1101/2020.03.20.20037325.
Roberts M, Driggs D, Thorpe M et al (2021) Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat Mach Intell 3:199–217
Pu J, Leader J, Bandos A et al (2021) Automated quantification of COVID-19 severity and progression using chest CT images. Eur Radiol 31:436–446
Xiao L, Li P, Sun P, et al (2020) Development and validation of a deep learning-based model using computed tomography imaging for predicting disease severity of coronavirus disease 2019. Front Bioeng Biotechnol 8
Adadi A, Berrada M (2018) Peeking inside the black-box: a survey on explainable artificial intelligence (XAI). IEEE Access 6:52138–52160
Arrieta A, Díaz-Rodríguez N, Ser J et al (2020) Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inform Fusion 58:82–115
Jin C, Chen W, Cao Y et al (2020) Development and evaluation of an artificial intelligence system for COVID-19 diagnosis. Nat Commun 11:5088
Wang S, Liu Z, Rong Y et al (2019) Deep learning provides a new computed tomography-based prognostic biomarker for recurrence prediction in high-grade serous ovarian cancer. Radiother Oncol 132:171–177
Morozov S, Andreychenko, A, Blokhin I, Vladzymyrskyy A, Gelezhe P, Gombolevskiy V, Gonchar A, Ledikhova N, Pavlov N, Chernina V (2020) MosMedData: chest CT scans with COVID-19 related findings. arXiv. https://doi.org/10.48550/arXiv.2005.06465
Peng Y, Tang Y, Lee S et al COVID-19-CT-CXR: A freely accessible and weakly labeled chest X-Ray and CT image collection on COVID-19 from biomedical literature. In: IEEE transactions on big data, vol. 7, no 1, pp 3–12. https://doi.org/10.1109/TBDATA.2020.3035935
Hofmanninger J, Prayer F, Pan J et al (2020) Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem. Eur Radiol Exp 4:50. https://doi.org/10.1186/s41747-020-00173-2
Zhang X, Li X, Feng Y (2015) A medical image segmentation algorithm based on bi-directional region growing. Optik 126(20):2398–2404. https://doi.org/10.1016/j.ijleo.2015.06.011
Hinton G, Srivastava N, Krizhevsky A et al (2012) Improving neural networks by preventing co-adaptation of feature detectors[J]. Comput Sci 3(4):212–223.
Chen L, Papandreou G, Schroff F, Adam H (2017) Rethinking atrous convolution for semantic image segmentation. ArXiv, abs/1706.05587.
Wang B, Jin S, Yan Q et al (2021) AI-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical AI system. Appl Soft Comput 98:106897
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest to report regarding this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, J., Li, Y., Guo, L. et al. Machine learning techniques for CT imaging diagnosis of novel coronavirus pneumonia: a review. Neural Comput & Applic 36, 181–199 (2024). https://doi.org/10.1007/s00521-022-07709-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-022-07709-0