Abstract
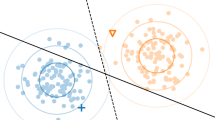
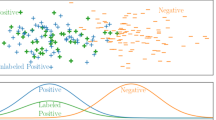
In several real-world node label prediction problems on graphs, in fields ranging from computational biology to World Wide Web analysis, nodes can be partitioned into categories different from the classes to be predicted, on the basis of their characteristics or their common properties. Such partitions may provide further information about node classification that classical machine learning algorithms do not take into account. We introduce a novel family of parametric Hopfield networks (m-category Hopfield networks) and a novel algorithm (Hopfield multi-category—HoMCat), designed to appropriately exploit the presence of property-based partitions of nodes into multiple categories. Moreover, the proposed model adopts a cost-sensitive learning strategy to prevent the remarkable decay in performance usually observed when instance labels are unbalanced, that is, when one class of labels is highly underrepresented than the other one. We validate the proposed model on both synthetic and real-world data, in the context of multi-species function prediction, where the classes to be predicted are the Gene Ontology terms and the categories the different species in the multi-species protein network. We carried out an intensive experimental validation, which on the one hand compares HoMCat with several state-of-the-art graph-based algorithms, and on the other hand reveals that exploiting meaningful prior partitions of input data can substantially improve classification performances.







Similar content being viewed by others
References
Ashburner M et al (2000) Gene ontology: tool for the unification of biology. Gene ontology consortium. Nat Genet 25(1):25–29
Atencia M, Joya G, Sandoval F (2004) Parametric identification of robotic systems with stable time-varying Hopfield networks. Neural Comput Appl 13(4):270–280. doi:10.1007/s00521-004-0421-4
Attwood TK, Bradley P, Flower DR, Gaulton A, Maudling N, Mitchell A, Moulton G, Nordle A, Paine K, Taylor P et al (2003) Prints and its automatic supplement, preprints. Nucl Acids Res 31(1):400–402
Azran A (2007) The rendezvous algorithm: multi- class semi-supervised learning with Markov random walks. In: Proceedings of the 24th international conference on machine learning (ICML)
Bairoch A, Apweiler R (1997) the SWISS-PROT protein sequence data bank and its supplement TrEMBL. Nucl Acids Res 25(1):31–36
Bengio Y, Delalleau O, Le Roux N (2006) Label propagation and quadratic criterion. In: Chapelle O, Scholkopf B, Zien A (eds) Semi supervised learning. MIT Press, Cambridge, pp 193–216
Bertoni A, Frasca M, Valentini G (2011) Cosnet: a cost sensitive neural network for semi-supervised learning in graphs. In: ECML/PKDD (1), Lecture Notes in Computer Science, vol 6911, pp 219–234. Springer
Bhagat S, Cormode G, Muthukrishnan S (2011) Node classification in social networks. CoRR abs/1101.3291
Bogdanov P, Singh AK (2010) Molecular function prediction using neighborhood features. IEEE/ACM Trans Comput Biol Bioinform 7:208–217
Brent R (1973) Algorithms for minimization without derivatives. Prentice-Hall, New Jersey
Chaudhari G, Avadhanula V, Sarawagi S (2014) A few good predictions: selective node labeling in a social network. In: Proceedings of the 7th ACM international conference on web search and data mining, WSDM ’14, pp 353–362. ACM, New York. doi:10.1145/2556195.2556241
Chen RM, Huang YM (2001) Multiprocessor task assignment with fuzzy Hopfield neural network clustering technique. Neural Comput Appl 10(1):12–21. doi:10.1007/s005210170013
Cheng Z, Caverlee J, Lee K (2010) You are where you tweet: a content-based approach to geo-locating twitter users. In: Proceedings of the 19th ACM international conference on information and knowledge management., CIKM ’10ACM, New York, pp 759–768
Chua HN, Sung WK, Wong L (2006) Exploiting indirect neighbours and topological weight to predict protein function from protein-protein interactions. Bioinformatics 22:1623–1630
Deng M, Chen T, Sun F (2004) An integrated probabilistic model for functional prediction of proteins. J Comput Biol 11:463–475
Elkan C (2001) The foundations of cost-sensitive learning. In: Proceedings of the seventeenth international joint conference on artificial intelligence, pp 973–978
Erdem MH, Ozturk Y (1996) A new family of multivalued networks. Neural Netw 9(6):979–989
Ertoz L, Steinbach M, Kumar V (2002) A new shared nearest neighbor clustering algorithm and its applications. In: Workshop on clustering high dimensional data and its applications at 2nd SIAM international conference on data mining
Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R et al (2006) Pfam: clans, web tools and services. Nucl Acids Res 34(suppl 1):D247–D251
Frasca M (2015) Automated gene function prediction through gene multifunctionality in biological networks. Neurocomputing. doi: 10.1016/j.neucom.2015.04.007. http://www.sciencedirect.com/science/article/pii/S0925231215004142. In press
Frasca M, Bertoni A et al (2013) A neural network algorithm for semi-supervised node label learning from unbalanced data. Neural Netw 43:84–98
Frasca M, Pavesi G (2013) A neural network based algorithm for gene expression prediction from chromatin structure. In: IJCNN, pp 1–8. IEEE
Gough J, Karplus K, Hughey R, Chothia C (2001) Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol 313(4):903–919
Guyon I, Cawley G, Dror G (eds) (2011) Hands-on pattern recognition: challenges in machine learning, challenges in machine learning, vol 1. Microtome Publishing, Brookline
Hebb DO (2002) The organization of behavior: a neuropsychological theory. Lawrence Erlbaum Associates Inc, US, Mahwah. http://www.loc.gov/catdir/enhancements/fy0659/2002018867-d.html
Hopfield J (1982) Neural networks and physical systems with emergent collective computational abilities. Proc Natl Acad Sci USA 79:2554–2558
Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJ (2006) The PROSITE database. Nucl Acids Res 34(suppl 1):D227–D230
Jarvis RA, Patrick EA (1973) Clustering using a similarity measure based on shared near neighbors. IEEE Trans Comput 22(11):1025–1034
Karaoz U et al (2004) Whole-genome annotation by using evidence integration in functional-linkage networks. Proc Natl Acad Sci USA 101:2888–2893
Kohler S, Bauer S, Horn D, Robinson P (2008) Walking the interactome for prioritization of candidate disease genes. Am J Human Genet 82(4):948–958
Kordos M, Duch W (2008) Variable step search algorithm for feedforward networks. Neurocomputing 71(13–15):2470–2480. doi:10.1016/j.neucom.2008.02.019
Lan L et al (2013) MS-kNN: protein function prediction by integrating multiple data sources. BMC Bioinformatics 14(Suppl 3:S8)
Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P (2006) Smart 5: domains in the context of genomes and networks. Nucl Acids Res 34(suppl 1):D257–D260
Ling C, Sheng V (2010) Class imbalance problem. In: Sammut C, Webb G (eds) Encyclopedia of machine learning, Springer, US, pp 171–171. doi:10.1007/978-0-387-30164-8_110
Ling C, Sheng V (2010) Cost-sensitive learning. In: Sammut C, Webb G (eds) Encyclopedia of machine learning, Springer, US, pp. 231–235. doi:10.1007/978-0-387-30164-8_181
Lovász L (1996) Random walks on graphs: a survey. In: Miklós D, Sós VT, Szőnyi T (eds) Combinatorics, Paul Erdős is eighty, vol 2. János Bolyai Mathematical Society, Budapest, pp 353–398
Ma J (1999) The object perceptron learning algorithm on generalised Hopfield networks for associative memory. Neural Comput Appl 8(1):25–32. doi:10.1007/s005210050004
Marcotte E, Pellegrini M, Thompson M, Yeates T, Eisenberg D (1999) A combined algorithm for genome-wide prediction of protein function. Nature 402:83–86
Mayer ML, Hieter P (2000) Protein networks-built by association. Nat Biotechnol 18(12):1242–3
Mérida-Casermeiro E, Galán-Marín G, Muñoz Pérez J (2001) An efficient multivalued Hopfield network for the traveling salesman problem. Neural Process Lett 14(3):203–216. doi:10.1023/A:1012751230791
Mesiti M, Re M, Valentini G (2014) Think globally and solve locally: secondary memory-based network learning for automated multi-species function prediction. Giga Sci 3:5. doi:10.1186/2047-217X-3-5
Mislove A, Viswanath B, Gummadi KP, Druschel P (2010) You are who you know: inferring user profiles in online social networks. In: Proceedings of the third ACM international conference on web search and data mining, WSDM ’10. ACM, New York, pp 251–260. doi:10.1145/1718487.1718519
Mostafavi S, Morris Q (2010) Fast integration of heterogeneous data sources for predicting gene function with limited annotation. Bioinformatics 26(14):1759–1765
Mostafavi S, Ray D, Farley DW, et al (2008) GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol 9(Suppl 1), S4+
Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, Copley R et al (2007) New developments in the InterPro database. Nucl Acids Res 35(suppl 1):D224–D228
Muller J, Szklarczyk D, Julien P, Letunic I, Roth A, Kuhn M, Powell S, von Mering C, Doerks T, Jensen LJ et al (2010) eggnog v2. 0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucl Acids Res 38(suppl 1):D190–D195
Murali TM, Wu CJ, Kasif S (2006) The art of gene function prediction. Nat Biotechnol 24(12):1474–1475. doi:10.1038/nbt1206-1474
Muruganantham G, Bhakat RS (2013) A review of impulse buying behavior. Int J Mark Stud 5(3):p149
Nabieva E, Jim K, Agarwal A, Chazelle B, Singh M (2005) Whole-proteome prediction of protein function via graph-theoretic analysis of interaction maps. Bioinformatics 21(S1):302–310
Neagu D, Palade V (2003) A neuro-fuzzy approach for functional genomics data interpretation and analysis. Neural Comput Appl 12(3–4):153–159. doi:10.1007/s00521-003-0388-6
Nie F, Xiang S, Liu Y, Zhang C (2010) A general graph-based semi-supervised learning with novel class discovery. Neural Comput Appl 19(4):549–555. doi:10.1007/s00521-009-0305-8
Pena-Castillo L, Tasan M, Myers C et al (2008) A critical assessment of Mus musculus gene function prediction using integrated genomic evidence. Genome Biol 9:S1
Radivojac P et al (2013) A large-scale evaluation of computational protein function prediction. Nat Methods 10(3):221–227
Re M, Mesiti M, Valentini G (2012) A fast ranking algorithm for predicting gene functions in biomolecular networks. IEEE/ACM Trans Comput Biol Bioinform 9(6):1812–1818. doi:10.1109/TCBB.2012.114
Re M, Valentini G (2012) Cancer module genes ranking using kernelized score functions. BMC Bioinform 13(Suppl 14/S3). doi:10.1186/1471-2105-13-S14-S3. http://www.biomedcentral.com/bmcbioinformatics/supplements/13/S14/S3
Salavati AH, Kumar KR, Shokrollahi A (2013) A non-binary associative memory with exponential pattern retrieval capacity and iterative learning: Extended Results. CoRR abs/1302.1156
Schwikowski B, Uetz P, Fields S (2000) A network of protein-protein interactions in yeast. Nat Biotechnol 18(12):1257–1261
Silva I, Moody G, Scott DJ, Celi LA, Mark RG (2012) Predicting in-hospital mortality of icu patients: the physionet/computing in cardiology challenge 2012. Comput Cardiol 39:245–248. http://www.biomedsearch.com/nih/Predicting-In-Hospital-Mortality-ICU/24678516.html
Szummer M, Jaakkola T (2001) Partially labeled classification with Markov random walks. In: Advances in neural information processing systems (NIPS) 14:945–952. MIT Press
Tsuda K, Shin H, Scholkopf B (2005) Fast protein classification with multiple networks. Bioinformatics 21(Suppl 2):ii59–ii65
Valentini G, Paccanaro A, Caniza H, Romero A, Re M (2014) An extensive analysis of disease-gene associations using network integration and fast kernel-based gene prioritization methods. Artif Intell Med 61(2):63–78. doi:10.1016/j.artmed.2014.03.003
Vazquez A, Flammini A, Maritan A, Vespignani A (2003) Global protein function prediction from protein-protein interaction networks. Nat Biotechnol 21:697–700
Wilcoxon F (1945) Individual comparisons by ranking methods. Biometrics 1:80–83
Wolfram Research Inc: Mathematica (2012) http://www.wolfram.com/mathematica/. Version 9.0
Wong AK, Park CY, Greene CS, Bongo LA, Guan Y, Troyanskaya OG (2012) Imp: a multi-species functional genomics portal for integration, visualization and prediction of protein functions and networks. Nucl Acids Res 40(W1):W484–W490
Xue H, Chen S (2011) Glocalization pursuit support vector machine. Neural Comput Appl 20(7):1043–1053. doi:10.1007/s00521-010-0448-7
Yoon K, Kwek S (2007) A data reduction approach for resolving the imbalanced data issue in functional genomics. Neural Comput Appl 16(3):295–306. doi:10.1007/s00521-007-0089-7
Youngs N, Penfold-Brown D, Drew K, Shasha D, Bonneau R (2013) Parametric Bayesian priors and better choice of negative examples improve protein function prediction. Bioinformatics 29(9):btt110–1198. doi:10.1093/bioinformatics/btt110
Zhou D et al (2004) Learning with local and global consistency. In: Thrun S, Saul L, Schölkopf B (eds) Advances in neural information processing systems 16:321–328. MIT Press. http://papers.nips.cc/paper/2506-learning-with-local-and-global-consistency
Zhu X, Ghahramani Z, Lafferty J (2003) Semi-supervised learning using Gaussian fields and harmonic functions. In. In ICML, pp 912–919
Zurada JM, Cloete I, van der Poel E (1996) Generalized Hopfield networks for associative memories with multi-valued stable states. Neurocomputing 13(24):135–149
Acknowledgments
Most of the main ideas behind this paper have been discussed with Alberto Bertoni. We would like to dedicate this paper to Alberto, who recently departed his life.
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix 1: Convergence proof
The following fact adapts the convergence proof for Hopfield networks with neuron activation values \(\{\sin \alpha , -\cos \alpha \}\) [21] to the case with \(m\,\ge\,2\) categories of neurons \(V_1, V_2,\ldots , V_m\), each with a different couple of activation values \(\{\sin \alpha _k, -\cos \alpha _k\}\), for neurons belonging to category \(V_k\).
Fact 5
A m-Category Hopfield Network \({\fancyscript{H}} = \langle \,\varvec{W}, \varvec{b}, \alpha _1, \ldots , \alpha _m, \varvec{\lambda }\rangle\) with neurons \(V=\{ 1, 2,\ldots , n\}\) and asynchronous dynamics (1), starting from any given network state, eventually reaches a stable state at a local minimum of the energy function.
Proof
We observe that the energy (2) is equivalent to the following
During the iteration \(t+1\), each unit i is selected and updated according to the update rule (1). To simplify the notation we set \(x'_i = x_i(t+1)\) and \(x_i = x_i(t)\). If the unit i does not change its state, then the energy of the system does not change as well, and we set \(x'_i = x_i\) . Otherwise, the network reaches a new global state \(\varvec{x}' = (x_1, \ldots , x'_i, \ldots , x_n)\) having energy \(E(\varvec{x}'\)). Since by definition \(W_{ii} = 0\), the difference between \(E(\varvec{x}'\)) and \(E(\varvec{x}\)) is given by all terms in the summation (11) which contain \(x'_i\) and \(x_i\), that is,
where \(B_i = \sum _{j=1}^n{ W_{ij}x_j } - \lambda _i\). The factor \(\frac{1}{2}\) disappears from the computation because the terms \(W_{ij} x_i x_j\) appear twice in the double summation in (11). If \(B_i > 0\), it means that \(x'_i = \sin \alpha _{b_i}\) and \(x_i = -\cos \alpha _{b_i}\) (see 1); since \(0\le \alpha _{b_i}< \frac{\pi }{2}\), \((x'_i - x_i) > 0\) and \(E(\varvec{x}') - E(\varvec{x}) < 0\). If \(B_i < 0\), it means that \(x'_i = -\cos \alpha _{b_i}\) and \(x_i = \sin \alpha _{b_i}\) and again \(E(\varvec{x}') - E(\varvec{x}) < 0\). If \(B_i\) = 0, the energy value does not change after the update. Hence, the energy (11) is a monotonically decreasing function. Moreover, since the connections \(W_{ij}\) are nonnegative, the energy E is lower bounded by the value
and the dynamics is guaranteed to converge to a fixed point, corresponding to a local minimum of the energy.\(\square\)
Appendix 2: Generative process for synthetic data
We provide here detailed information about the generative process of the two families of experiments on synthetic data described in Sect. 5.1. The generator used in the experiments is available from the authors upon request.
The main prerequisite to perform the first family of experiments is the generation of separable patterns. For this purpose we decided to work in the \(\Delta\) space, where each node \(i\in V\) is mapped in a 2m-dimensional point \(\Delta (i)\equiv \{\Delta ^+_1(i),\Delta ^-_1(i),\ldots ,\Delta ^+_m(i),\Delta ^-_m(i)\}\) where
and, according to the notation used throughout the paper, \(V_k\) is the k-th category, \(S^+\) (resp. \(S^-\)) the set of positive (resp. negative) labeled instances, and \(W_{ij}\) the weight connecting nodes i and j.
Namely, after having fixed a labeling imbalance \(\epsilon\), we randomly generated a set of n points \(\Delta (i)\) in the unitary hypercube, partitioned them into m categories according to their Euclidean similarity in the \(\Delta\) space. Then, we drew \(\alpha _k\) uniformly for each category k, and computed the threshold \(\lambda\) guaranteeing the correct labeling imbalance, i.e., satisfying the following equation:
where \({\rm {HS}}\) is the Heaviside step function, i.e., \({\rm {HS}}(x)=1\) if \(x\ge 0\), 0 otherwise.
We run the proposed learning algorithm starting with m categories, and subsequently evaluated its performance on a lower number \(\widetilde{m}< m\) of them. Although other strategies could be adopted, the decision of working directly in the \(\Delta\) space was motivated by the need of exerting more control on the construction of separable instances. We select the number \(\widetilde{m}\) of categories to be investigated as a divisor of m. For instance, in case \(m=4\), by choosing \(\widetilde{m}=2\) we may easily consider the reduced \(\Delta\) space obtained by associating with node i the point \(\{\Delta _1^+(i)+\Delta _2^+(i),\ \Delta _1^-(i)+\Delta _2^-(i),\ \Delta _3^+(i)+\Delta _4^+(i),\ \Delta _3^-(i)+\Delta _4^-(i)\}\) which, in turn, can be coupled with two parameters (\(\alpha _1\) and \(\alpha _2)\) out of four. Of course, in order to preserve the concept of category, this aggregation should be performed by merging together the most similar categories, where similarity is again defined in terms of Euclidean metric in the \(\Delta\) space.
Given the set of nodes V, partitioned into positives \(V_+\) and negatives \(V_-\), the vector \(\varvec{b}\) partitioning V in m disjoint categories and the corresponding parameters \((\alpha _1, \ldots , \alpha _m, \lambda )\), we compute the initial labeling \({\tilde{\varvec{x}}}\) as follows:
Then, by exploiting the associative memory functionality of the Hopfield network, we generated the weight matrix \(\varvec{W}\) through the naive Hebbian rule [25] in order to store the pattern \({\tilde{\varvec{x}}}\), i.e., \({\tilde{\varvec{x}}}\) becomes a fixed point of the dynamics [26]. Moreover, to make the convergence to \({\tilde{\varvec{x}}}\) as much complex as possible and hence to amplify the fixed point perturbations, we learned the weights \(\varvec{W}\) in order to store also other \(\mu\) randomly generated patterns with the same label imbalance \(\epsilon = \frac{|V_+|}{|V_-|}\), where \(\mu \simeq 0.13|V|\) is the capacity of the network when trained with the naive Hebbian rule. Then, after having hidden the labels for a subset \(U \subset V\) chosen according to the fixed \(\frac{|S|}{|V|}\) ratio, for each value of the standard deviation \(\sigma\) modulating the injected noise, we run the network restricted to U till convergence and finally we measure the perturbation of the attained stable state with respect to \({\tilde{\varvec{x}}}_u\).
Rights and permissions
About this article
Cite this article
Frasca, M., Bassis, S. & Valentini, G. Learning node labels with multi-category Hopfield networks. Neural Comput & Applic 27, 1677–1692 (2016). https://doi.org/10.1007/s00521-015-1965-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-015-1965-1