Abstract
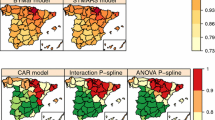
One of the main objectives in disease mapping is the identification of temporal trends and the production of a series of smoothed maps from which spatial patterns of mortality risks can be monitored over time. When studying rare diseases, conditional autoregressive models have been commonly used for smoothing risks. In this work, a P-spline ANOVA type model is used instead. The model is anisotropic and explicitly considers different smooth terms for space, time, and space-time interaction avoiding, in addition, model identifiability problems. The mean squared error of the log-risk predictor is derived accounting for the variability associated to the estimation of the smoothing parameters. The procedure is illustrated analyzing Spanish prostate cancer mortality data in the period 1975–2008.





Similar content being viewed by others
References
Baade PD, Youlden DR, Krnjacki LJ (2009) International epidemiology of prostate cancer: geographical distribution and secular trends. Mol Nutr Food Res 53:171–184
Besag J, York J, Mollié A (1991) Bayesian image restoration with two applications in spatial statistics, Ann Inst Stat Math 43:1–59
Bernardinelli L, Clayton D, Pascutto C, Montomoli C, Ghislandi M, Songini M (1995) Bayesian analysis of space-time variation in disease risk. Stat Med 14:2433–2443
Breslow NE, Clayton DG (1993) Approximate inference in generalized linear mixed models. J Am Stat Assoc 88: 9–25
Cabanes A, Vidal E, Aragonés N, Pérez-Gómez B, Pollán M, Lope V, López-Abente G (2010) Cancer mortality trends in Spain: 1980–2007. Ann Oncol 21(3):14–20
Cabanes A, Pérez-Gómez B, Aragonés N, Pollán M, López-Abente G (2009) The situation of cancer in Spain, 1975–2006 (In Spanish). Instituto de Salud Carlos III. Madrid
Cayuela A, Rodriguez-Dominguez S, Vigil-Martin E, Barrero-Candau BR (2008) Cambios recientes en la mortalidad por cáncer de próstata en España: estudio de tendencias en el periodo 1991–2005. Actas Urol Esp 32:184–189
Chen Z (1993) Fitting multivariate regression functions by interactions spline models. J R Stat Soc B 55: 473-491
Das K, Jiang J, Rao JNK (2004) Mean squared error of empirical predictor. Ann Stat 32:818–840
Dean CB, Ugarte MD, Militino AF (2004) Penalized quasi-likelihood with spatially correlated data. Comput Stat Data Anal 45:235–248
de Boor C (1978) A practical guide to splines. Springer, Berlin
Eilers PHC, Currie ID, Durbán M (2006) Fast and compact smoothing on large multidimensional grids. Comput Stat Data Anal 50:61–76
Eilers PHC, Marx BD (1996) Flexible smoothing with B-splines and penalties. Stat Sci 11:89–121
Escaramís G, Carrasco JL, Ascaso C (2008) Detection of significant disease risks using a spatial conditional autoregressive model. Biometrics 64:1043–1053
Ferlay J, Bray F, Pisani P, Parkin DM. GLOBOCAN 2002 (2004) Cancer incidence, mortality and prevalence worldwide. IARC CancerBase No. 5. version 2.0. IARC, Lyon
Gu C (2002) Smoothing spline ANOVA models. Springer, New York
Harville DA (1977) Maximum likelihood approaches to variance component estimation and to related problems. J Am Stat Assoc 72:320–340
Kackar R and Harville DA (1984) Approximations for standard errors of estimators of fixed and random effects in mixed linear models. J Am Stat Assoc 79:853–862
Knorr-Held L (2000) Bayesian modelling of inseparable space-time variation in disease risk. Stat Med 19:2555–2567
Knorr-Held L, Besag J (1998) Modelling risk from a disease in time and space. Stat Med 17:2045–2060
Larrañaga N, Galceran J, Ardanaz E, Navarro C, Sánchez MJ, Pastor-Berriuso R. (2010) Prostate cancer incidence trends in Spain before and during the prostate-specific antigen era: impact on mortality. Ann Oncol 21(3):83–89
Lee DJ (2010) Smoothing mixed models for spatial and spatio-temporal data Ph-D Thesis, Carlos III University, Madrid. http://e-archivo.uc3m.es/browse?type=author&value=Lee+Hwang%2C+Dae-Jin
Lee DJ, Durbán M (2011) P-spline ANOVA-type interaction models for spatio-temporal smoothing. Stat Model 11:49–69
MacNab YC and Dean CB (2001) Autoregressive spatial smoothing and temporal spline smoothing for mapping rates. Biometrics 57:949–956
MacNab YC and Dean CB (2002) Spatio-temporal modelling of rates for the construction of disease maps. Stat Med 21:347–358
Martínez-Beneito MA, López-Quílez A, Botella-Rocamora P (2008) An autoregressive approach to spatio-temporal disease mapping. Stat Med 27:2874–2889
Opsomer JD, Claeskens G, Ranalli MG, Kauermann G, Breidt FJ (2008) Non-parametric small area estimation using penalized spline regression. J R Stat Soc B 70:265–286
Parkin DM, Whelan P, Ferlay J, Storm H (2005) Cancer incidence in five continents Vols. I–VIII. IARC Cancer Base No. 7. IARC, Lyon
Prasad NGN, Rao JNK (1990) The estimation of mean squared error of small area estimators. J Am Stat Assoc 85:163–171
R Development Core Team (2011) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL: http://www.R-project.org/
Ruppert D, Wand MP, Carroll RJ (2003) Semiparametric regression Cambridge series in statistical and probabilistic mathematics. Cambridge University Press, New York
Ruppert D, Wand MP, Carroll RJ (2009) Semiparametric regression during 2003–2007. Electron J Stat 3:1193–1256
Schall R (1991) Estimation in generalized linear models with random effects. Biometrika 78:719-727
Singh BB, Shukla GK, Kundu D (2005) Spatio-temporal models in small area estimation. Surv Methodol 31:183–195
Tiefelsdorf M (2007) Controlling for migration effects in ecological disease mapping of prostate cancer. Stoch Environ Res Risk Asses 21:615–624
Ugarte MD (2009) Mortality. In: Kattan MW (ed) Encyclopedia of medical decision making, vol. 2. SAGE, Thousand Oaks, pp 788–792
Ugarte MD, Goicoa T, Militino AF (2010) Spatio-temporal modelling of mortality risks using penalized splines. Environmetrics 21:270–289
Ugarte MD, Militino AF, Goicoa T (2008) Prediction error estimators in empirical Bayes disease mapping. Environmetrics 19:287–300
Waller LA, Carlin BP, Xia H, Gelfand AE (1997) Hierarchical spatio-temporal mapping of disease rates. J Am Stat Assoc 92:607–617
Wood SN (2006) Low-Rank scale-invariant tensor product smooths for generalized additive mixed models. Biometrics 62:1025–1036
Acknowledgements
This research has been supported by the Spanish Ministry of Science and Innovation, project MTM2008-03085, and project MTM2011-22664, which is co-funded by FEDER. The authors would like to thank to Marina Pollán from the National Epidemiology Center (area of Environmental Epidemiology and Cancer) for providing the data.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
In this section, detailed expressions for \({{\partial{\bf G}}\over {\partial\lambda_j}}\) and \({{\partial{\bf V}^{-1}}\over {\partial\lambda_j}}\) are provided.
In this model there are six variance components corresponding to the smoothing parameters of the ANOVA type model
and then
with λ j denoting the jth component of λ.
Rights and permissions
About this article
Cite this article
Ugarte, M.D., Goicoa, T., Etxeberria, J. et al. A P-spline ANOVA type model in space-time disease mapping. Stoch Environ Res Risk Assess 26, 835–845 (2012). https://doi.org/10.1007/s00477-012-0570-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00477-012-0570-4