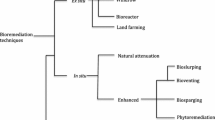
Abstract
Axenic microbial applications in the open environment are unrealistic and may not be always practically viable. Therefore, it is important to use mixed microbial cultures and their interactions with the microbiome in the targeted ecosystem to perform robust functions towards their sustainability in harsh environmental conditions. Emerging pollutants like phthalates and hydrocarbons that are toxic to several aquatic and terrestrial life forms in the water bodies and lands are an alarming situation. The present review explores the possibility of devising an inclusive eco-friendly strategy like microbiome engineering which proves to be a unique and crucial technology involving the power of microbial communication through quorum sensing. This review discusses the interspecies and intra-species communications between different microbial groups with their respective environments. Moreover, this review also envisages the efforts for designing the next level of microbiome-host engineering concept (MHEC). The focus of the review also extended toward using omics and metabolic network analysis-based tools for effective microbiome engineering. These approaches might be quite helpful in the future to understand such microbial interactions but it will be challenging to implement in the real environment to get the desired functions. Finally, the review also discusses multiple approaches for the bioremediation of toxic chemicals from the soil environment.


Similar content being viewed by others
References
Nai C, Meyer V (2018) From axenic to mixed cultures: technological advances accelerating a paradigm shift in microbiology. Trends Microbiol 26(6):538–554
Bassler BL, Losick R (2006) Bacterially speaking. Cell 125(2):237–246
Stenuit B, Agathos SN (2015) Deciphering microbial community robustness through synthetic ecology and molecular systems synecology. Curr Opin Biotechnol 33:305–317
Tsoi R, Wu F, Zhang C, Bewick S, Karig D, You L (2018) Metabolic division of labor in microbial systems. Proc Natl Acad Sci 115(10):2526–2531
Tsoi R, Dai Z, You L (2019) Emerging strategies for engineering microbial communities. Biotechnol Adv 37(6):107372
Xu P (2020) Dynamics of microbial competition, commensalism, and cooperation and its implications for coculture and microbiome engineering. Biotechnol Bioeng 118(1):199–209
Galitskaya P, Biktasheva L, Blagodatsky S, Selivanovskaya S (2021) Response of bacterial and fungal communities to high petroleum pollution in different soils. Sci Rep 11(1):1–18
Dsikowitzky L, Schwarzbauer J (2015) Hexa (methoxymethyl) melamine: an emerging contaminant in German rivers. Water Environ Res 87(5):461–469
Zhu F, Doyle E, Zhu C, Zhou D, Gu C, Gao J (2020) Metagenomic analysis exploring microbial assemblages and functional genes potentially involved in di (2-ethylhexyl) phthalate degradation in soil. Sci Total Environ 715:137037
Tian Z, Zhao H, Peter KT, Gonzalez M, Wetzel J, Wu C, Kolodziej EP (2021) A ubiquitous tire rubber–derived chemical induces acute mortality in coho salmon. Science 371(6525):185–189
Albright MB, Louca S, Winkler DE, Feeser KL, Haig SJ, Whiteson KL, Dunbar J (2021) Solutions in microbiome engineering: prioritizing barriers to organism establishment. ISME J 16(2):331–338
Lee ED, Aurand ER, Friedman DC, Engineering Biology Research Consortium Microbiomes Roadmapping Working Group (2020) Engineering Microbiomes—Looking Ahead. ACS Synth Biol 9(12):3181–3183
Graham EB, Knelman JE, Schindlbacher A, Siciliano S, Breulmann M, Yannarell A, Nemergut DR (2016) Microbes as engines of ecosystem function: when does community structure enhance predictions of ecosystem processes? Front Microbiol 7:214
Antwis RE, Griffiths SM, Harrison XA, Aranega-Bou P, Arce A, Bettridge AS, Sutherland WJ (2017) Fifty important research questions in microbial ecology. FEMS Microbiol Ecol 93(5):fix044
Fierer N (2017) Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol 15(10):579–590
Shetty SA, Kuipers B, Atashgahi S, Aalvink S, Smidt H, de Vos WM (2022) Inter-species metabolic interactions in an in-vitro minimal human gut microbiome of Core bacteria. NPJ Biofilms Microbiomes 8(1):1–13
Postma-Blaauw MB, De Goede RGM, Bloem J, Faber JH, Brussaard L (2010) Soil biota community structure and abundance under agricultural intensification and extensification. Ecology 91:460–473
Verbruggen E, Röling WFM, Gamper HA, Kowalchuk GA, Verhoef HA, van der Heijden MGA (2010) Positive effects of organic farming on below-ground mutualists: large-scale comparison of mycorrhizal fungal communities in agricultural soils. New Phytol 186:968–979
Waters CM, Bassler BL (2005) Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346
Ng WL, Bassler BL (2009) Bacterial quorum-sensing network architectures. Annu Rev Genet 43:197–222
Prescott RD, Decho AW (2020) Flexibility and adaptability of quorum sensing in nature. Trends Microbiol 28(6):436–444
Whiteley M, Diggle SP, Greenberg EP (2017) Progress in and promise of bacterial quorum sensing research. Nature 551(7680):313–320
Hawver LA, Jung SA, Ng WL (2016) Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol Rev 40(5):738–752
Schauder S, Shokat K, Surette MG, Bassler BL (2001) The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol 41(2):463–476
Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM (2002) Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415(6871):545–549
Bassler B, Vogel J (2013) Bacterial regulatory mechanisms: the gene and beyond. Curr Opin Microbiol 16(2):109–111
Williams P, Winzer K, Chan WC, Camara M (2007) Look who’s talking: communication and quorum sensing in the bacterial world. Philos Trans R Soc B Biol Sci 362(1483):1119–1134
Cárcamo-Oyarce G, Lumjiaktase P, Kümmerli R, Eberl L (2015) Quorum sensing triggers the stochastic escape of individual cells from Pseudomonas putida biofilms. Nat Commun 6(1):1–9
Fuqua WC, Winans SC, Greenberg EP (1994) Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 176(2):269–275
Schuster M, Joseph Sexton D, Diggle SP, Peter Greenberg E (2013) Acyl-homoserine lactone quorum sensing: from evolution to application. Annu Rev Microbiol 67:43–63
Papenfort K, Bassler BL (2016) Quorum sensing signal–response systems in gram-negative bacteria. Nat Rev Microbiol 14(9):576–588
Kiers ET, Rousseau RA, West SA, Denison RF (2003) Host sanctions and the legume–rhizobium mutualism. Nature 425(6953):78–81
Dandekar AA, Chugani S, Greenberg EP (2012) Bacterial quorum sensing and metabolic incentives to cooperate. Science 338(6104):264–266
Subramoni S, Venturi V (2009) LuxR-family ‘solos’: bachelor sensors/regulators of signalling molecules. Microbiology 155(5):1377–1385
Hudaiberdiev S, Choudhary KS, Vera Alvarez R, Gelencsér Z, Ligeti B, Lamba D, Pongor S (2015) Census of solo LuxR genes in prokaryotic genomes. Front Cell Infect Microbiol 5:20
Fuqua C, Greenberg EP (1998) Self-perception in bacteria: quorum sensing with acylated homoserine lactones. Curr Opin Microbiol 1(2):183–189
Ryan RP, An SQ, Allan JH, McCarthy Y, Dow JM (2015) The DSF family of cell–cell signals: an expanding class of bacterial virulence regulators. PLoS Pathog 11(7):e1004986
Subramoni S, Florez Salcedo DV, Suarez-Moreno ZR (2015) A bioinformatic survey of distribution, conservation, and probable functions of LuxR solo regulators in bacteria. Front Cell Infect Microbiol 5:16
Ahmer BM, Van Reeuwijk J, Timmers CD, Valentine PJ, Heffron F (1998) Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J Bacteriol 180(5):1185–1193
Michael B, Smith JN, Swift S, Heffron F, Ahmer BM (2001) SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J Bacteriol 183(19):5733–5742
Smith JN, Ahmer BM (2003) Detection of other microbial species by Salmonella: expression of the SdiA regulon. J Bacteriol 185(4):1357–1366
Rajput A, Kumar M (2017) In silico analyses of conservational, functional and phylogenetic distribution of the LuxI and LuxR homologs in Gram-positive bacteria. Sci Rep 7(1):1–13
Rajput A, Kumar M (2017) Computational exploration of putative LuxR Solos in archaea and their functional implications in quorum sensing. Front Microbiol 8:798
Biswa P, Doble M (2013) Production of acylated homoserine lactone by Gram-positive bacteria isolated from marine water. FEMS Microbiol Lett 343(1):34–41
Banerjee S, Zhao C, Kirkby CA, Coggins S, Zhao S, Bissett A, Richardson AE (2021) Microbial interkingdom associations across soil depths reveal network connectivity and keystone taxa linked to soil fine-fraction carbon content. Agr Ecosyst Environ 320:107559
TeamR RC (2017). A language and environment for statistical computing Vienna. Austria R Foundation for Statistical Computing
Bahram M, Hildebrand F, Forslund SK, Anderson JL, Soudzilovskaia NA, Bodegom PM, Bork P (2018) Structure and function of the global topsoil microbiome. Nature 560(7717):233–237
Deveau A, Bonito G, Uehling J, Paoletti M, Becker M, Bindschedler S, Wick LY (2018) Bacterial–fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol Rev 42(3):335–352
Worrich A, Wick LY, Banitz T (2018) Ecology of contaminant biotransformation in the mycosphere: role of transport processes. Adv Appl Microbiol 104:93–133
Furuno S, Foss S, Wild E, Jones KC, Semple KT, Harms H, Wick LY (2012) Mycelia promote active transport and spatial dispersion of polycyclic aromatic hydrocarbons. Environ Sci Technol 46(10):5463–5470
Ferguson BA, Dreisbach TA, Parks CG, Filip GM, Schmitt CL (2003) Coarse-scale population structure of pathogenic Armillaria species in a mixed-conifer forest in the Blue Mountains of northeast Oregon. Can J For Res 33(4):612–623
Ritz K, Young IM (2004) Interactions between soil structure and fungi. Mycologist 18(2):52–59
Wongsuk T, Pumeesat P, Luplertlop N (2016) Fungal quorum sensing molecules: role in fungal morphogenesis and pathogenicity. J Basic Microbiol 56(5):440–447
Cugini C, Calfee MW, Farrow JM III, Morales DK, Pesci EC, Hogan DA (2007) Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol Microbiol 65(4):896–906
Stanley CE, Stöckli M, van Swaay D, Sabotič J, Kallio PT, Künzler M, Aebi M (2014) Probing bacterial–fungal interactions at the single cell level. Integr Biol 6(10):935–945
Sztajer H, Szafranski SP, Tomasch J, Reck M, Nimtz M, Rohde M, Wagner-Döbler I (2014) Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J 8(11):2256–2271
Dixon EF, Hall RA (2015) Noisy neighbourhoods: quorum sensing in fungal–polymicrobial infections. Cell Microbiol 17(10):1431–1441
Fourie R, Ells R, Swart CW, Sebolai OM, Albertyn J, Pohl CH (2016) Candida albicans and Pseudomonas aeruginosa interaction, with focus on the role of eicosanoids. Front Physiol 7:64
Trejo-Hernández A, Andrade-Domínguez A, Hernández M, Encarnacion S (2014) Interspecies competition triggers virulence and mutability in Candida albicans–Pseudomonas aeruginosa mixed biofilms. ISME J 8(10):1974–1988
Wagg C, Schlaeppi K, Banerjee S, Kuramae EE, van der Heijden MG (2019) Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat Commun 10(1):1–10
Kohlmeier S, Smits TH, Ford RM, Keel C, Harms H, Wick LY (2005) Taking the fungal highway: mobilization of pollutant-degrading bacteria by fungi. Environ Sci Technol 39(12):4640–4646
Lindahl BD, Ihrmark K, Boberg J, Trumbore SE, Högberg P, Stenlid J, Finlay RD (2007) Spatial separation of litter decomposition and mycorrhizal nitrogen uptake in a boreal forest. New Phytol 173(3):611–620
Van Der Heijden MG, Bruin SD, Luckerhoff L, Van Logtestijn RS, Schlaeppi K (2016) A widespread plant-fungal-bacterial symbiosis promotes plant biodiversity, plant nutrition and seedling recruitment. ISME J 10(2):389–399
Durán P, Thiergart T, Garrido-Oter R, Agler M, Kemen E, Schulze-Lefert P, Hacquard S (2018) Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 175(4):973–983
Blaustein RA, Lorca GL, Meyer JL, Gonzalez CF, Teplitski M (2017) Defining the core citrus leaf- and root-associated microbiota: factors associated with community structure and implications for managing huanglongbing (citrus greening) disease. Appl Environ Microbiol 83:e00210-e217
Hamonts K, Trivedi P, Garg A, Janitz C, Grinyer J, Holford P, Singh BK (2018) Field study reveals core plant microbiota and relative importance of their drivers. Environ Microbiol 20(1):124–140
Xu J, Zhang Y, Zhang P, Trivedi P, Riera N, Wang Y, Wang N (2018) The structure and function of the global citrus rhizosphere microbiome. Nat Commun 9(1):1–10
Zhang R, Vivanco JM, Shen Q (2017) The unseen rhizosphere root–soil–microbe interactions for crop production. Curr Opin Microbiol 37:8–14
Liu H, Brettell LE, Qiu Z, Singh BK (2020) Microbiome-mediated stress resistance in plants. Trends Plant Sci 25(8):733–743
Carrión VJ, Perez-Jaramillo J, Cordovez V, Tracanna V, De Hollander M, Ruiz-Buck D, Raaijmakers JM (2019) Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 366(6465):606–612
Hassan S, Mathesius U (2012) The role of flavonoids in root–rhizosphere signalling: opportunities and challenges for improving plant–microbe interactions. J Exp Bot 63(9):3429–3444
Neal AL, Ahmad S, Gordon-Weeks R, Ton J (2012) Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS One 7(4):e35498
Kudjordjie EN, Sapkota R, Steffensen SK, Fomsgaard IS, Nicolaisen M (2019) Maize synthesized benzoxazinoids affect the host associated microbiome. Microbiome 7(1):1–17
Hu L, Robert CA, Cadot S, Zhang XI, Ye M, Li B, Erb M (2018) Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat Commun 9(1):1–13
McSpadden Gardener BB, Weller DM (2001) Changes in populations of rhizosphere bacteria associated with take-all disease of wheat. Appl Environ Microbiol 67(10):4414–4425
Lombardi N, Vitale S, Turrà D, Reverberi M, Fanelli C, Vinale F, Lorito M (2018) Root exudates of stressed plants stimulate and attract Trichoderma soil fungi. Mol Plant Microbe Interact 31(10):982–994
Lanoue A, Burlat V, Henkes GJ, Koch I, Schurr U, Röse US (2010) De novo biosynthesis of defense root exudates in response to Fusarium attack in barley. New Phytol 185(2):577–588
Stringlis IA, Yu K, Feussner K, de Jonge R, Van Bentum S, Van Verk MC, Pieterse CM (2018) MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc Natl Acad Sci 115(22):E5213–E5222
Huang AC, Jiang T, Liu YX, Bai YC, Reed J, Qu B, Osbourn A (2019) A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 364(6440):eaau6389
Van Deynze A, Zamora P, Delaux PM, Heitmann C, Jayaraman D, Rajasekar S, Bennett AB (2018) Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLoS Biol 16(8):e2006352
Schulz-Bohm K, Gerards S, Hundscheid M, Melenhorst J, de Boer W, Garbeva P (2018) Calling from distance: attraction of soil bacteria by plant root volatiles. ISME J 12(5):1252–1262
Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP (2001) Quorum-sensing in gram-negative bacteria. FEMS Microbiol Rev 25(4):365–404
D’Angelo-Picard C, Faure D, Penot I, Dessaux Y (2005) Diversity of N-acyl homoserine lactone-producing and-degrading bacteria in soil and tobacco rhizosphere. Environ Microbiol 7(11):1796–1808
Dong YH, Wang LH, Xu JL, Zhang HB, Zhang XF, Zhang LH (2001) Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411(6839):813–817
Trivedi P, Mattupalli C, Eversole K, Leach JE (2021) Enabling sustainable agriculture through understanding and enhancement of microbiomes. New Phytol 230(6):2129–2147
de Souza RSC, Okura VK, Armanhi JSL, Jorrín B, Lozano N, Da Silva MJ, Arruda P (2016) Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome. Sci Rep 6(1):1–15
Simonin M, Dasilva C, Terzi V, Ngonkeu ELM, Diouf D, Kane A, Bena G, Moulin L (2020) Influence of plant genotype and soil on the wheat rhizosphere microbiome: evidences for a core microbiome across eight African and European soils. FEMS Microbiol Ecol 96:067
Singha LP, Pandey P (2021) Rhizosphere assisted bioengineering approaches for the mitigation of petroleum hydrocarbons contamination in soil. Crit Rev Biotechnol 41(5):749–766
Sun Q, Li A, Li M, Hou B (2015) Effect of pH on biodiesel production and the microbial structure of glucose-fed activated sludge. Int Biodeterior Biodegradation 104:224–230
Peng M, Zi X, Wang Q (2015) Bacterial community diversity of oil-contaminated soils assessed by high throughput sequencing of 16S rRNA genes. Int J Environ Res Public Health 12(10):12002–12015
Jiao S, Liu Z, Lin Y, Yang J, Chen W, Wei G (2016) Bacterial communities in oil contaminated soils: biogeography and co-occurrence patterns. Soil Biol Biochem 98:64–73
Subedi R, Taupe N, Ikoyi I, Bertora C, Zavattaro L, Schmalenberger A, Grignani C (2016) Chemically and biologically-mediated fertilizing value of manure-derived biochar. Sci Total Environ 550:924–933
Auti AM, Narwade NP, Deshpande NM, Dhotre DP (2019) Microbiome and imputed metagenome study of crude and refined petroleum-oil-contaminated soils: Potential for hydrocarbon degradation and plant-growth promotion. J Biosci 44(5):1–16
Towell MG, Bellarby J, Paton GI, Coulon F, Pollard SJ, Semple KT (2011) Mineralisation of target hydrocarbons in three contaminated soils from former refinery facilities. Environ Pollut 159(2):515
Liu Q, Tang J, Gao K, Gurav R, Giesy JP (2017) Aerobic degradation of crude oil by microorganisms in soils from four geographic regions of China. Sci Rep 7(1):1–12
Sutton NB, Maphosa F, Morillo JA, Abu Al-Soud W, Langenhoff AA, Grotenhuis T, Smidt H (2013) Impact of long-term diesel contamination on soil microbial community structure. Appl Environ Microbiol 79(2):619–630
Yang S, Wen X, Shi Y, Liebner S, Jin H, Perfumo A (2016) Hydrocarbon degraders establish at the costs of microbial richness, abundance and keystone taxa after crude oil contamination in permafrost environments. Sci Rep 6(1):1–13
Aislabie J, Saul DJ, Foght JM (2006) Bioremediation of hydrocarbon-contaminated polar soils. Extremophiles 10(3):171–179
Ma Y, Wang L, Shao Z (2006) Pseudomonas, the dominant polycyclic aromatic hydrocarbon-degrading bacteria isolated from Antarctic soils and the role of large plasmids in horizontal gene transfer. Environ Microbiol 8(3):455–465
Margesin R, Moertelmaier C, Mair J (2013) Low-temperature biodegradation of petroleum hydrocarbons (n-alkanes, phenol, anthracene, pyrene) by four actinobacterial strains. Int Biodeterior Biodegradation 84:185–191
Walker BH (1992) Biodiversity and ecological redundancy. Conserv biol 6(1):18–23
Yachi S, Loreau M (1999) Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc Natl Acad Sci 96(4):1463–1468
Loreau M (2004) Does functional redundancy exist? Oikos 104(3):606–611
Proulx SR, Promislow DE, Phillips PC (2005) Network thinking in ecology and evolution. Trends Ecol Evol 20(6):345–353
Vidal M, Cusick ME, Barabási AL (2011) Interactome networks and human disease. Cell 144(6):986–998
Layeghifard M, Hwang DM, Guttman DS (2017) Disentangling interactions in the microbiome: a network perspective. Trends Microbiol 25(3):217–228
Matchado MS, Lauber M, Reitmeier S, Kacprowski T, Baumbach J, Haller D, List M (2021) Network analysis methods for studying microbial communities: a mini review. Comput Struct Biotechnol J 19:2687–2698
Li X, Meng D, Li J, Yin H, Liu H, Liu X, Yan M (2017) Response of soil microbial communities and microbial interactions to long-term heavy metal contamination. Environ Pollut 231:908–917
Magnúsdóttir S, Heinken A, Kutt L, Ravcheev DA, Bauer E, Noronha A, Thiele I (2017) Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat Biotechnol 35(1):81–89
Dukovski I, Bajić D, Chacón JM, Quintin M, Vila JC, Sulheim S, Segrè D (2021) A metabolic modeling platform for the computation of microbial ecosystems in time and space (COMETS). Nat Protoc 16(11):5030–5082
Chan SHJ, Simons MN, Maranas CD (2017) SteadyCom: predicting microbial abundances while ensuring community stability. PLoS Comput Biol 13(5):e1005539
García-Jiménez B, García JL, Nogales J (2018) FLYCOP: metabolic modeling-based analysis and engineering microbial communities. Bioinformatics 34(17):i954–i963
Kehe J, Kulesa A, Ortiz A, Ackerman CM, Thakku SG, Sellers D, Blainey PC (2019) Massively parallel screening of synthetic microbial communities. Proc Natl Acad Sci 116(26):12804–12809
Tandogan N, Abadian PN, Epstein S, Aoi Y, Goluch ED (2014) Isolation of microorganisms using sub-micrometer constrictions. PLoS One 9(6):e101429
Grünberger A, Probst C, Helfrich S, Nanda A, Stute B, Wiechert W, Kohlheyer D (2015) Spatiotemporal microbial single-cell analysis using a high-throughput microfluidics cultivation platform. Cytometry A 87(12):1101–1115
Hansen RH, Timm AC, Timm CM, Bible AN, Morrell-Falvey JL, Pelletier DA, Retterer ST (2016) Stochastic assembly of bacteria in microwell arrays reveals the importance of confinement in community development. PLoS One 11(5):e0155080
Grünberger A, Schöler K, Probst C, Kornfeld G, Hardiman T, Wiechert W, Noack S (2017) Real-time monitoring of fungal growth and morphogenesis at single-cell resolution. Eng Life Sci 17(1):86–92
Aleklett K, Kiers ET, Ohlsson P, Shimizu TS, Caldas VE, Hammer EC (2018) Build your own soil: exploring microfluidics to create microbial habitat structures. ISME J 12(2):312–319
Kim HJ, Du W, Ismagilov RF (2011) Complex function by design using spatially pre-structured synthetic microbial communities: degradation of pentachlorophenol in the presence of Hg (II). Integr Biol 3(2):126–133
Burmeister A, Grünberger A (2020) Microfluidic cultivation and analysis tools for interaction studies of microbial co-cultures. Curr Opin Biotechnol 62:106–115
Nagy K, Ábrahám Á, Keymer JE, Galajda P (2018) Application of microfluidics in experimental ecology: the importance of being spatial. Front Microbiol 9:496
Mandáková T, Singh V, Krämer U, Lysak MA (2015) Genome structure of the heavy metal hyperaccumulator Noccaea caerulescens and its stability on metalliferous and nonmetalliferous soils. Plant Physiol 169(1):674–689
Auguy F, Fahr M, Moulin P, El Mzibri M, Smouni A, Filali-Maltouf A, Doumas P (2016) Transcriptome changes in Hirschfeldia incana in response to lead exposure. Front Plant Sci 6:1231
Briskine RV, Paape T, Shimizu-Inatsugi R, Nishiyama T, Akama S, Sese J, Shimizu KK (2017) Genome assembly and annotation of Arabidopsis halleri, a model for heavy metal hyperaccumulation and evolutionary ecology. Mol Ecol Resour 17(5):1025–1036
Basharat Z, Novo LA, Yasmin A (2018) Genome editing weds CRISPR: what is in it for phytoremediation? Plants 7(3):51
Jaiswal S, Singh DK, Shukla P (2019) Gene editing and systems biology tools for pesticide bioremediation: a review. Front Microbiol 10:87
Yadav R, Kumar V, Baweja M, Shukla P (2018) Gene editing and genetic engineering approaches for advanced probiotics: a review. Crit Rev Food Sci Nutr 58(10):1735–1746
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339(6121):819–823
Tyagi S, Kumar R, Das A, Won SY, Shukla P (2020) CRISPR-Cas9 system: a genome-editing tool with endless possibilities. J Biotechnol 319:36–53
Andersen MM, Landes X, Xiang W, Anyshchenko A, Falhof J, Østerberg JT, Palmgren MG (2015) Feasibility of new breeding techniques for organic farming. Trends Plant Sci 20(7):426–434
Ali Z, Abul-Faraj A, Li L, Ghosh N, Piatek M, Mahjoub A, Mahfouz MM (2015) Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol Plant 8(8):1288–1291
Chen A, Huang Y (2020) Acyl homoserine lactone based quorum sensing affects phenanthrene removal by Novosphingobium pentaromativorans US6-1 through altering cell surface properties. Int Biodeterior Biodegrad 147:104841
McCarty NS, Ledesma-Amaro R (2019) Synthetic biology tools to engineer microbial communities for biotechnology. Trends Biotechnol 37(2):181–197
Suenaga H, Watanabe T, Sato M, Ngadiman, Furukawa K (2002) Alteration of regiospecificity in biphenyl dioxygenase by active-site engineering. J Bacteriol 184(13):3682–3688
Chebrou H, Hurtubise Y, Barriault D, Sylvestre M (1999) Heterologous expression and characterization of the purified oxygenase component of Rhodococcus globerulus P6 biphenyl dioxygenase and of chimeras derived from it. J Bacteriol 181(16):4805–4811
Wang W, Hou J, Zheng N, Wang X, Zhang J (2019) Keeping our eyes on CRISPR: the “Atlas” of gene editing. Cell Biol Toxicol 35(4):285–288
Li R, Liu C, Zhao R, Wang L, Chen L, Yu W, Shen L (2019) CRISPR/Cas9-Mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol 19(1):1–13
Tang L, Yang F, He X, Xie H, Liu X, Fu J, Gu F (2019) Efficient cleavage resolves PAM preferences of CRISPR-Cas in human cells. Cell Regen 8(2):44–50
Rubin BE, Diamond S, Cress BF, Crits-Christoph A, Lou YC, Borges AL, Doudna JA (2022) Species-and site-specific genome editing in complex bacterial communities. Nat Microbiol 7(1):34–47
Zengler K, Hofmockel K, Baliga NS, Behie SW, Bernstein HC, Brown JB, Northen TR (2019) EcoFABs: advancing microbiome science through standardized fabricated ecosystems. Nat Methods 16(7):567–571
Ke J, Wang B, Yoshikuni Y (2021) Microbiome engineering: synthetic biology of plant-associated microbiomes in sustainable agriculture. Trends Biotechnol 39(3):244–261
Silva-Castro GA, Uad I, Gónzalez-López J, Fandiño CG, Toledo FL, Calvo C (2012) Application of selected microbial consortia combined with inorganic and oleophilic fertilizers to recuperate oil-polluted soil using land farming technology. Clean Technol Environ Policy 14(4):719–726
Ali SS, Mustafa AM, Kornaros M, Manni A, Sun J, Khalil MA (2020) Construction of novel microbial consortia CS-5 and BC-4 valued for the degradation of catalpa sawdust and chlorophenols simultaneously with enhancing methane production. Biores Technol 301:122720
Shen T, Pi Y, Bao M, Xu N, Li Y, Lu J (2015) Biodegradation of different petroleum hydrocarbons by free and immobilized microbial consortia. Environ Sci Process Impacts 17(12):2022–2033
Zafra G, Absalón ÁE, Anducho-Reyes MÁ, Fernandez FJ, Cortés-Espinosa DV (2017) Construction of PAH-degrading mixed microbial consortia by induced selection in soil. Chemosphere 172:120–126
Li F, Liu Y, Wang D, Zhang C, Yang Z, Lu S, Wang Y (2018) Biodegradation of di-(2-ethylhexyl) phthalate by a halotolerant consortium LF. PLoS One 13(10):e0204324
Yang J, Guo C, Liu S, Liu W, Wang H, Dang Z, Lu G (2018) Characterization of a di-n-butyl phthalate-degrading bacterial consortium and its application in contaminated soil. Environ Sci Pollut Res 25(18):17645–17653
Krainara S, Suraraksa B, Prommeenate P, Thayanukul P, Luepromchai E (2020) Enrichment and characterization of bacterial consortia for degrading 2-mercaptobenzothiazole in rubber industrial wastewater. J Hazard Mater 400:123291
Lu M, Jiang W, Gao Q, Zhang M, Hong Q (2020) Degradation of dibutyl phthalate (DBP) by a bacterial consortium and characterization of two novel esterases capable of hydrolyzing PAEs sequentially. Ecotoxicol Environ Saf 195:110517
Shariati S, Pourbabaee AA, Alikhani HA, Rezaei KA (2021) Biodegradation of DEHP by a new native consortium An6 (Gordonia sp. and Pseudomonas sp.) adapted with phthalates, isolated from a natural strongly polluted wetland. Environ Technol Innov 24:101936
Zhang L, Qiu X, Huang L, Xu J, Wang W, Li Z, Tang H (2021) Microbial degradation of multiple PAHs by a microbial consortium and its application on contaminated wastewater. J Hazard Mater 419:126524
Vejan P, Abdullah R, Khadiran T, Ismail S, Nasrulhaq Boyce A (2016) Role of plant growth promoting rhizobacteria in agricultural sustainability—a review. Molecules 21(5):573
Qiu Z, Egidi E, Liu H, Kaur S, Singh BK (2019) New frontiers in agriculture productivity: optimised microbial inoculants and in situ microbiome engineering. Biotechnol Adv 37(6):107371
Li DS, Feng JQ, Liu YF, Zhou L, Liu JF, Gu JD, Yang SZ (2019) Enrichment and immobilization of oil-degrading microbial consortium on different sorbents for bioremediation testing under simulated aquatic and soil conditions. Appl Environ Biotechnol 5(1):1–11
Li J, Guo C, Liao C, Zhang M, Liang X, Lu G, Dang Z (2016) A bio-hybrid material for adsorption and degradation of phenanthrene: bacteria immobilized on sawdust coated with a silica layer. RSC Adv 6(109):107189–107199
Partovinia A, Rasekh B (2018) Review of the immobilized microbial cell systems for bioremediation of petroleum hydrocarbons polluted environments. Crit Rev Environ Sci Technol 48(1):1–38
Zhen-Yu W, Ying XU, Hao-Yun W, Jian Z, Dong-Mei G, Feng-Min L, Xing B (2012) Biodegradation of crude oil in contaminated soils by free and immobilized microorganisms. Pedosphere 22(5):717–725
Parvatha Reddy P (2012) Bio-priming of Seeds: Recent Advances in Crop Protection. Springer, New Delhi
Niu B, Paulson JN, Zheng X, Kolter R (2017) Simplified and representative bacterial community of maize roots. Proc Natl Acad Sci 114(12):E2450–E2459
Pill WG, Collins CM, Goldberger B, Gregory N (2009) Responses of non-primed or primed seeds of ‘Marketmore 76’cucumber (Cucumis sativus L) slurry coated with Trichoderma species to planting in growth media infested with Pythium aphanidermatum. Sci Hortic 121(1):54–62
Mendes R, Garbeva P, Raaijmakers JM (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37(5):634–663
Yadav RS, Singh V, Pal S, Meena SK, Meena VS, Sarma BK, Rakshit A (2018) Seed bio-priming of baby corn emerged as a viable strategy for reducing mineral fertilizer use and increasing productivity. Sci Hortic 241:93–99
Halmer P (2000) Commercial seed treatment technology. Seed technology and its biological basis. Academic Press, Sheffield, England, pp 257–286
Goswami AP, Vishunavat K, Mohan C, Ravi S (2017) Effect of seed coating, storage periods and storage containers on soybean (Glycine max (L.) Merrill) seed quality under ambient conditions. J Appl Nat Sci 9(1):598–602
Mei J, Wang W, Peng S, Nie L (2017) Seed pelleting with calcium peroxide improves crop establishment of direct-seeded rice under waterlogging conditions. Sci Rep 7(1):1–12
Tu L, He Y, Shan C, Wu Z (2016) Preparation of microencapsulated Bacillus subtilis SL-13 seed coating agents and their effects on the growth of cotton seedlings. BioMed Res Int 2016:1–7
He Y, Wu Z, Tu L, Han Y, Zhang G, Li C (2015) Encapsulation and characterization of slow-release microbial fertilizer from the composites of bentonite and alginate. Appl Clay Sci 109:68–75
Acknowledgements
LPS acknowledges financial support received from SERB in form of National Post-Doctoral Fellowship (NPDF) through Grant No. PDF/2021/000223. PS acknowledges, the Lab Infrastructure grant by BHU, Varanasi (F(C)/XVIII-Spl.Fund/Misc/Infrastructure/Instt.Sc/2019-2020/10290) and BTiS—Sub-Distributed Information Centre, funded by DBT, Govt. of India at the School of Biotechnology, Banaras Hindu University, Varanasi, India.
Funding
Science and Engineering Research Board, PDF/2021/000223.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Singha, L.P., Shukla, P. Microbiome engineering for bioremediation of emerging pollutants. Bioprocess Biosyst Eng 46, 323–339 (2023). https://doi.org/10.1007/s00449-022-02777-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-022-02777-x