Abstract
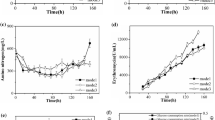
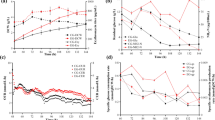
In this paper, several different fermentation experiments were designed to address whether modulating glucose and propanol feeds could benefit the production level of erythromycin during pilot plant (30 L) fermentation. Results showed that glucose feed rate (determined by a set high or low culture pH) had no effect on erythromycin production, indicating that glucose was not the limiting factor for erythromycin biosynthesis under these conditions. It was found that decreasing glucose feed could stimulate the consumption of propanol, and the high erythromycin production (12.49 ± 0.50 mg ml−1) was achieved by controlling the feed rates of glucose and propanol. The quantitative metabolic flux analysis disclosed that high propanol consumption increased the pool size of propionyl-CoA (~2.147 mmol g−1 day−1) and methylmalonyl-CoA (~1.708 mmolg−1 day−1). It was also found that 45–77 % of the propanol went into the TCA cycle which strengthened the conclusion that blocking the propionate pathway to TCA cycle could lead to a significant increase in erythromycin production in carbohydrate-based media (Reeves et al. Ind Microbiol Biotechnol 7:600–609, 2006). In addition, the results also suggested that a relative low intracellular ATP level resulting from low glucose feed did not limit the erythromycin biosynthesis, and a relatively high NADPH should be beneficial for erythromycin biosynthesis.






Similar content being viewed by others
References
Mironov VA, Sergienko OV, Nastasyak IN, Danilenko VN (2004) Biogenesis and regulation of biosynthesis of erythromycins in Saccharopolysplra erythraea. Appl Biochem Microbiol 6:611–624
Chng C, Lum AM, Vroom JA, Kao CM (2008) A key developmental regulator controls the synthesis of the antibiotic erythromycin in Saccharopolyspora erythraea. PNAS 32:11346–11351
Reeves AR, Cernota WH, Brikun IA, Wesley RK, Weber JM (2004) Engineering precursor flow for increased erythromycin production in Aeromicrobium erythreum. Metab Eng 4:300–312
Reeves AR, Brikun IA, Cernota WH, Leach BI, Gonzalez MC, Weber JM (2006) Effects of methylmalonyl-CoA mutase gene knockouts on erythromycin production in carbohydrate-based and oil-based fermentations of Saccharopolyspora erythraea. Ind Microbiol Biotechnol 7:600–609
Wang Y, Wang YG, Chu J, Zhuang YP, Zhang LX, Zhang SL (2007) Improved production of erythromycin A by expression of a heterologous gene encoding S-adenosylmethionine synthetase. Appl Microbiol Biotechnol 4:837–842
Brunker P, Minas W, Kallio PT, Bailey JE (1998) Genetic engineering of an industrial strain of Saccharopolyspora erythraea for stable expression of the Vitreoscilla haemoglobin gene(vhb). Microbiol 9:2441–2448
Chen Y, Deng W, Wu JQ, Qian JC, Chu J, Zhuang YP, Zhang SL, Liu W (2008) Genetic modulation of the overexpression of tailoring genes eryK and eryG leading to the improvement of erythromycin A purity and production in Saccharopolyspora erythraea fermentation. Appl Environ Microb 6:1820–1828
Elmahdi I, Baganz F, Dixon K, Harrop T, Sugden D, Lye GJ (2003) pH control in microwell fermentations of S. erythraea CA340: influence on biomass growth kinetics and erythromycin biosynthesis. Biochem Eng J 3:299–310
Zou X, Chen CF, Hang HF, Chu J, Zhuang YP, Zhang SL (2010) Response surface methodology for optimization of the erythromycin production by fed-batch fermentation using an inexpensive biological nitrogen source. Chem Biochem Eng Q 1:95–100
Zou X, Hang HF, Chu J, Zhuang YP, Zhang SL (2009) Oxygen uptake rate optimization with nitrogen regulation for erythromycin production and scale-up from 50 L to 372 m (3) scale. Bioresour Technol 3:1406–1412
Zou X, Hang HF, Chu J, Zhuang YP, Zhang SL (2009) Enhancement of erythromycin A production with feeding available nitrogen sources in erythromycin biosynthesis phase. Bioresour Technol 13:3358–3365
Staunton J, Wilkinson B (1997) Biosynthesis of erythromycin and rapamycin. Chem Rev 7:2611–2630
Reeves AR, Brikun IA, Cernota WH, Leach BI, Gonzalez MC, Weber JM (2007) Engineering of the methylmalonyl-CoA metabolite node of Saccharopolyspora erythraea for increased erythromycin production. Metab Eng 3:293–303
Weber JM, Cernota WH, Gonzalez MC, Leach BI, Reeves AR, Wesley RK (2012) An erythromycin process improvement using the diethyl methylmalonate-responsive (Dmr) phenotype of the Saccharopolyspora erythraea mutB strain. Appl Microbiol Biotechnol 4:1575–1583
Chan YA, Podevels AM, Kevany B M, Thomas GM (2009) Biosynthesis of polyketide synthase extender units. Nat Prod Rep 1:90–114
Bojanowsa KR, Ruczaj Z, Korszynka DS, Rafalski A (1973) Limiting reaction in activation of acyl units in biosynthesis of macrolide antibiotics. Antimicrob Agents CH 2:162–167
Bermúdez O, Padilla P, Huitrón C, Flores ME (1998) Influence of carbon and nitrogen source on synthesis of NADP+-isocitrate dehydrogenase, methylmalonyl-coenzyme A mutase, and methylmalonyl-coenzyme A decarboxylase in Saccharopolyspora erythraea CA340. FEMS Microbiol Lett 1:77–82
Trilli A, Crossley MV, Kontakou M (1987) Relation between growth rate and erythromycin production in Streptomyces erythraeus. Biotechnol Lett 11:765–770
Potvin J, Peringer P (1994) Ammonium regulation in Saccharopolyspora erythraea. Part II: growth and antibiotic production. Biotechnol Lett 1:69–74
Liang JG, Chu XH, Xiong ZQ, Chu J, Wang YH (2011) Oxygen uptake rate regulation during cell growth phase for improving avermectin B1a batch fermentation on a pilot scale (2 m3). Microbiol Biotechnol 27:2639–2644
Textor S, Wendisch VF, Graaf AD, Müller U, Linder MI, Linder D, Buckel W (1997) Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch Microbiol 5:428–436
Murli S, Kennedy J, Dayem LC, Carney JR, Kealey JT (2003) Metabolic engineering of Escherichia coli for improved 6-deoxyerythronolide B production. Ind Microbiol Biotechnol 8:500–509
McPherson M, Khosla C, Cane DE (1998) Erythromycin biosynthesis: the β-ketoreductase domains catalyze the stereospecific transfer of the 4-pro-S hydride of NADPH. J Am Chem Soc 13:3267–3268
Cane DE, Kudo F, Kinoshita K, Khosla C (2002) Precursor-directed biosynthesis: biochemical basis of the remarkable selectivity of the erythromycin polyketide synthase toward unsaturated triketides. Chem and Biol 1:131–142
Li JH, Yang YM, Huang MZ, Li L, Zhang XC, Wang YH, Zhuang YP, Zhang SL (2010) Quantitative metabolic flux analysis revealed uneconomical utilization of ATP and NADPH in Acremonium chrysogenum fed with soybean oil. Bioprocess Biosyst Eng 9:1119-1129
Lambalot RH, Cane DE (1995) Overproduction and characterization of the erythromycin C-12 hydroxylase EryK. Biochem 34:1858–1866
Acknowledgments
This work was financially supported by a grant from the Major State Basic Research Development Program of China (973 Program, No. 2012CB721006), National Natural Science Foundation of China (No. 21276081), the National Scientific and Technological Major Special Project (Significant Creation of New drugs, No. 2011ZX09203-001-03), and Research Fund for the Doctoral Program of Higher Education of China (No. 20110074110015).
Author information
Authors and Affiliations
Corresponding authors
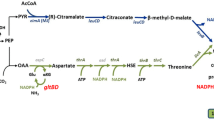
Appendix: Biochemical reactions in the metabolic model
Appendix: Biochemical reactions in the metabolic model
-
(a)
Uptake reactions
-
1.
Glucose + ATP → glucose-6-phosphate
-
2.
Palmitic acid + ATP + 8CoASH → 8acetyl-CoA + 7FADH + 7NADH + AMP + 2Pi
-
3.
Stearic acid + ATP + 9CoASH → 9acetyl-CoA + 8FADH + 8NADH + AMP + Pi
-
4.
Arachidic acid + ATP + 10CoASH → 10acetyl-CoA + 9FADH + 9NADH + AMP + Pi
-
5.
Oleic acid + ATP + 9CoASH → 9acetyl-CoA + 7FADH + 8NADH + AMP + Pi
-
6.
Linoleic acid + ATP + 9CoASH + NADPH → 9acetyl-CoA + 6FADH + 8NADH + AMP + 2Pi
-
7.
Linolenic acid +ATP + 9CoASH + NADPH → 9acetyl-CoA + FADH + 8NADH + AMP + Pi
-
8.
C117.17H218.37O15.86(soybean oil) → C3H8O3(glycerol) + C114.17H210.37O12.86(fatty acid)
-
9.
Fatty acid + 6.4ATP + 50.1CoA + 3.9NADPH → 50.1acetyl-CoA + 40.9FADH + 50.7NADH + 6.4AMP
-
10.
Glycerol → glycerate-3-phosphate + NADH
-
11.
Propanol → Propionyl-CoA + 2NADH
-
1.
-
(b)
Embden–Meyerhoff–Parnas pathway
-
1.
Glucose-6-phosphate → Fructose-6-phosphate
-
2.
Fructose-6-phosphate + ATP → 2glycerate-3-phosphate + ADP + Pi
-
3.
Glycerate-3-phosphate → Pyruvate + ATP + NADH
-
1.
-
(c)
TCA cycle
-
1.
Pyruvate + CoASH → acetyl-CoA + ATP + NADH + CO2
-
2.
Acetyl-CoA + oxaloacetic acid → citrate
-
3.
Citrate → α-ketoglutaric acid + NADH + CO2
-
4.
α-Ketoglutaric acid + CoASH → succinyl-CoA + NADH + CO2
-
5.
Succinyl-CoA + ADP → succinate + ATP + NADH
-
6.
Succinyl-CoA → oxaloacetic acid
-
1.
-
(d)
Pentose–phosphate cycle
-
1.
Glucose-6-phosphate → Ribulose-5-phosphate + 2NADPH + CO2
-
2.
3Ribulose-5-phosphate → 2Fructose-6-phosphate + glycerate-3-phosphate
-
1.
-
(e)
Mitochondrial reactions
-
1.
Pyruvate + ATP + CO2 → oxaloacetic acid
-
2.
Pyruvate → organic acids
-
3.
NADH + 0.5O2 → 2.5ATP
-
4.
FADH + 0.5O2 → 1.5ATP
-
1.
-
(f)
Erythromycin pathway
-
1.
Succinyl-CoA → methylmalonyl-CoA
-
2.
Propionyl-CoA + ATP + CO2 → methylmalonyl-CoA
-
3.
Propionyl-CoA + oxaloacetic acid → methylmalonyl-CoA + Pyruvate
-
4.
Propionyl-CoA + 6methylmalonyl-CoA + 2glucose + 9NADPH → erythromycin + 2NADH + 6CO2
-
1.
Rights and permissions
About this article
Cite this article
Chen, Y., Huang, M., Wang, Z. et al. Controlling the feed rate of glucose and propanol for the enhancement of erythromycin production and exploration of propanol metabolism fate by quantitative metabolic flux analysis. Bioprocess Biosyst Eng 36, 1445–1453 (2013). https://doi.org/10.1007/s00449-013-0883-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-013-0883-9