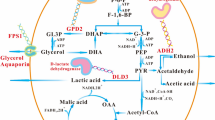
Abstract
In industrial process, yeast cells are exposed to ethanol stress that affects the cell growth and the productivity. Thus, investigating the intracellular state of yeast cells under high ethanol concentration is important. In this study, using DNA microarray analysis, we performed comprehensive expression profiling of two strains of Saccharomyces cerevisiae, i.e., the ethanol-adapted strain that shows active growth under the ethanol stress condition and its parental strain used as the control. By comparing the expression profiles of these two strains under the ethanol stress condition, we found that the genes related to ribosomal proteins were highly up-regulated in the ethanol-adapted strain. Further, genes related to ATP synthesis in mitochondria were suggested to be important for growth under ethanol stress. We expect that the results will provide a better understanding of ethanol tolerance of yeast.

Similar content being viewed by others
References
Attfield PV (1997) Stress tolerance: the key to effective strains of industrial baker’s yeast. Nat Biotechnol 15:1351–1357
Gibson BR, Lawrence SJ, Leclaire JP, Powell CD, Smart KA (2007) Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol Rev 31:535–569
Kajiwara S, Suga K, Sone H, Nakamura K (2000) Improved ethanol tolerance of Saccharomyces cerevisiae strains by increases in fatty acid unsaturation via metabolic engineering. Biotechnol Lett 22:1839–1843
Ogawa Y, Nitta A, Uchiyama H, Imamura T, Shimoi H, Ito K (2000) Tolerance mechanism of the ethanol-tolerant mutant of sake yeast. J Biosci Bioeng 90:313–320
Watanabe M, Tamura K, Magbanua JP, Takano K, Kitamoto K, Kitagaki H, Akao T, Shimoi H (2007) Elevated expression of genes under the control of stress response element (STRE) and Msn2p in an ethanol-tolerance sake yeast Kyokai no. 11. J Biosci Bioeng 104:163–170
Alexandre H, Ansanay-Galeote V, Dequin S, Blondin B (2001) Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett 498:98–110
Rossignol T, Dulau L, Julien A, Blondin B (2003) Genome-wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast 20:1369–1385
Hirasawa T, Yoshikawa K, Nakakura Y, Nagahisa K, Furusawa C, Katakura Y, Shimizu H, Shioya S (2007) Identification of target genes conferring ethanol stress tolerance to Saccharomyces cerevisiae based on DNA microarray data analysis. J Biotechnol 131:34–44
Pérez-Torrado R, Carrasco P, Aranda A, Gimeno-Alcañiz J, Pérez-Ortín JE, Matallana E, del Olmo ML (2002) Study of the first hours of microvinification by the use of osmotic stress-response genes as probes. Syst Appl Microbiol 25:153–161
Hirasawa T, Nakakura Y, Yoshikawa K, Ashitani K, Nagahisa K, Furusawa C, Katakura Y, Shimizu H, Shioya S (2006) Comparative analysis of transcriptional responses to saline stress in the laboratory and brewing strains of Saccharomyces cerevisiae with DNA microarray. Appl Microbiol Biotechnol 70:346–357
Dinh TN, Nagahisa K, Hirasawa T, Furusawa C, Shimizu H (2008) Adaptation of Saccharomyces cerevisiae cells to high ethanol concentration and changes in fatty acid composition of membrane and cell size. PLoS ONE 3:e2623
Winston F, Dollard C, Ricupero-Hovasse SL (1995) Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53–55
Köhrer K, Domdey H (1991) Preparation of high molecular weight RNA. Methods Enzymol 194:398–405
Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Güldener U, Mannhaupt G, Münsterkötter M, Mewes HW (2004) The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res 32:5539–5545
Costa V, Amorim MA, Reis E, Quintanilha A, Moradas-Ferreira P (1997) Mitochondrial superoxide dismutase is essential for ethanol tolerance of Saccharomyces cerevisiae in the post-diauxic phase. Microbiology 143:1649–1656
Martinez MJ, Roy S, Archuletta AB, Wentzell PD, Anna-Arriola SS, Rodriguez AL, Aragon AD, Quiñones GA, Allen C, Werner-Washburne M (2004) Genomic analysis of stationary-phase and exit in Saccharomyces cerevisiae: gene expression and identification of novel essential genes. Mol Biol Cell 15:5295–5305
Herman PK (2002) Stationary phase in yeast. Curr Opin Microbiol 5:602–607
Marks VD, Ho Sui SJ, Erasmus D, van der Merwe GK, Brumm J, Wasserman WW, Bryan J, van Vuuren HJ (2008) Dynamics of the yeast transcriptome during wine fermentation reveals a novel fermentation stress response. FEMS Yeast Res 8:35–52
Yoshikawa K, Tanaka T, Furusawa C, Nagahisa K, Hirasawa T, Shimizu H (2008) Comprehensive phenotypic analysis for identification of genes affecting growth under ethanol stress in Saccharomyces cereviciae. FEMS Yeast Res doi:10.1111/j.1567-1364.2008.00456.x
Kubota S, Takeo I, Kume K, Kanai M, Shitamukai A, Mizunuma M, Miyakawa T, Shimoi H, Iefuji H, Hirata D (2004) Effect of ethanol on cell growth of budding yeast: genes that are important for cell growth in the presence of ethanol. Biosci Biotechnol Biochem 68:968–972
van Voorst F, Houghton-Larsen J, Jønson L, Kielland-Brandt MC, Brandt A (2006) Genome-wide identification of genes required for growth of Saccharomyces cerevisiae under ethanol stress. Yeast 23:351–359
Fujita K, Matsuyama A, Kobayashi Y, Iwahashi H (2006) The genome-wide screening of yeast deletion mutants to identify the genes required for tolerance to ethanol and other alcohols. FEMS Yeast Res 6:744–750
Gibson BR, Boulton CA, Box WG, Graham NS, Lawrence SJ, Linforth RS, Smart KA (2008) Carbohydrate utilization and the larger yeast transcriptome during brewery fermentation. Yeast 25(8):549–562
Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G (2006) Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314:1565–1568
Chandler M, Stanley GA, Rogers P, Chambers P (2004) A genomic approach to defining the ethanol stress response in the yeast Saccharomyces cerevisiae. Ann Microbiol 54(4):427–454
Acknowledgments
This work was supported by Grant-in-Aids for Young Scientists (B) to CF and TH, respectively, from the Ministry of Education, Culture, Sports, Science and Technology of Japan. This work was also supported in part by “Global COE Program”, and “Special Coordination Funds for Promoting Science and Technology, Yuragi Project,” from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dinh, T.N., Nagahisa, K., Yoshikawa, K. et al. Analysis of adaptation to high ethanol concentration in Saccharomyces cerevisiae using DNA microarray. Bioprocess Biosyst Eng 32, 681–688 (2009). https://doi.org/10.1007/s00449-008-0292-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-008-0292-7