Abstract
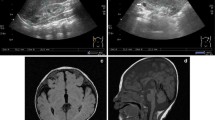
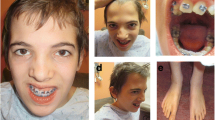
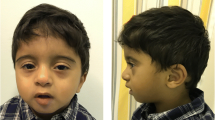
Cooks syndrome (CS) is an ultrarare limb malformation due to in tandem microduplications involving KCNJ2 and extending to the 5′ regulatory element of SOX9. To date, six CS families were resolved at the molecular level. Subsequent studies explored the evolutionary and pathological complexities of the SOX9-KCNJ2/Sox9-Kcnj2 locus, and suggested a key role for the formation of novel topologically associating domain (TAD) by inter-TAD duplications in causing CS. Here, we report a unique case of CS associated with a de novo 1;17 translocation affecting the KCNJ2 locus. On chromosome 17, the breakpoint mapped between KCNJ16 and KCNJ2, and combined with a ~ 5 kb deletion in the 5′ of KCNJ2. Based on available capture Hi-C data, the breakpoint on chromosome 17 separated KCNJ2 from a putative enhancer. Gene expression analysis demonstrated downregulation of KCNJ2 in both patient’s blood cells and cultured skin fibroblasts. Our findings suggest that a complex rearrangement falling in the 5′ of KCNJ2 may mimic the developmental consequences of in tandem duplications affecting the SOX9-KCNJ2/Sox9-Kcnj2 locus. This finding adds weight to the notion of an intricate role of gene regulatory regions and, presumably, the related three-dimensional chromatin structure in normal and abnormal human morphology.



Similar content being viewed by others
References
Castori M, Bottillo I, Morlino S, Barone C, Cascone P, Pediatric Craniofacial Malformation (PECRAM) Study Group, Grammatico P, Laino L (2016) Variability in a three-generation family with Pierre Robin sequence, acampomelic campomelic dysplasia, and intellectual disability due to a novel ~1 Mb deletion upstream of SOX9, and including KCNJ2 and KCNJ16. Birth Defects Res A Clin Mol Teratol 106(1):61–68. https://doi.org/10.1002/bdra.23463
Despang A, Schöpflin R, Franke M, Ali S, Jerković I, Paliou C, Chan WL, Timmermann B, Wittler L, Vingron M, Mundlos S, Ibrahim DM (2009) Functional dissection of the Sox9-Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat Genet 51(8):1263–1271. https://doi.org/10.1038/s41588-019-0466-z
Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485(7398):376–380. https://doi.org/10.1038/nature11082
ENCODE Project Consortium, Moore JE, Purcaro MJ, Pratt HE, Epstein CB, Shoresh N, Adrian J, Kawli T, Davis CA, Dobin A, Kaul R, Halow J, Van Nostrand EL, Freese P, Gorkin DU, Shen Y, He Y, Mackiewicz M, Pauli-Behn F, Williams BA, Mortazavi A, Keller CA, Zhang XO, Elhajjajy SI, Huey J, Dickel DE, Snetkova V, Wei X, Wang X, Rivera-Mulia JC, Rozowsky J, Zhang J, Chhetri SB, Zhang J, Victorsen A, White KP, Visel A, Yeo GW, Burge CB, Lécuyer E, Gilbert DM, Dekker J, Rinn J, Mendenhall EM, Ecker JR, Kellis M, Klein RJ, Noble WS, Kundaje A, Guigó R, Farnham PJ, Cherry JM, Myers RM, Ren B, Graveley BR, Gerstein MB, Pennacchio LA, Snyder MP, Bernstein BE, Wold B, Hardison RC, Gingeras TR, Stamatoyannopoulos JA, Weng Z (2020) Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583(7818):699–710. https://doi.org/10.1038/s41586-020-2493-4
Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schöpflin R, Kraft K, Kempfer R, Jerković I, Chan WL, Spielmann M, Timmermann B, Wittler L, Kurth I, Cambiaso P, Zuffardi O, Houge G, Lambie L, Brancati F, Pombo A, Vingron M, Spitz F, Mundlos S (2016) Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538(7624):265–269. https://doi.org/10.1038/nature19800
Gordon CT, Tan TY, Benko S, Fitzpatrick D, Lyonnet S, Farlie PG (2009) Long-range regulation at the SOX9 locus in development and disease. J Med Genet 46(10):649–656. https://doi.org/10.1136/jmg.2009.068361
Haraksingh RR, Snyder MP (2013) Impacts of variation in the human genome on gene regulation. J Mol Biol 425(21):3970–3977. https://doi.org/10.1016/j.jmb.2013.07.015
Haseeb A, Kc R, Angelozzi M, de Charleroy C, Rux D, Tower RJ, Yao L, Pellegrino da Silva R, Pacifici M, Qin L, Lefebvre V (2021) SOX9 keeps growth plates and articular cartilage healthy by inhibiting chondrocyte dedifferentiation/osteoblastic redifferentiation. Proc Natl Acad Sci USA 118(8):e2019152118. https://doi.org/10.1073/pnas.2019152118
Ibrahim DM, Mundlos S (2020) The role of 3D chromatin domains in gene regulation: a multi-facetted view on genome organization. Curr Opin Genet Dev 61:1–8. https://doi.org/10.1016/j.gde.2020.02.015
Jeong DU, Jiyeong Lee J, Lim KM (2020) Computational study to identify the effects of the KCNJ2 E299V mutation in cardiac pumping capacity. Comput Math Methods Med 31:7194275. https://doi.org/10.1155/2020/7194275
Jung I, Schmitt A, Diao Y, Lee AJ, Liu T, Yang D, Tan C, Eom J, Chan M, Chee S, Chiang Z, Kim C, Masliah E, Barr CL, Li B, Kuan S, Kim D, Ren B (2019) A compendium of promoter-centered long-range chromatin interactions in the human genome. Nat Genet 51(10):1442–1449. https://doi.org/10.1038/s41588-019-0494-8
Kurth I, Klopocki E, Stricker S, van Oosterwijk J, Vanek S, Altmann J, Santos HG, van Harssel JJ, de Ravel T, Wilkie AO, Gal A, Mundlos S (2009) Duplications of noncoding elements 5′ of SOX9 are associated with brachydactyly-anonychia. Nat Genet 41(8):862–863. https://doi.org/10.1038/ng0809-862
Liu M, Zhang X, Liu H, Shen Y (2020) A 17q24.3 duplication identified in a large Chinese family with brachydactyly-anonychia. Mol Genet Genomic Med. https://doi.org/10.1002/mgg3.1392
Lupiáñez DG, Spielmann M, Mundlos S (2016) Breaking TADs: how alterations of chromatin domains result in disease. Trends Genet 32(4):225–237. https://doi.org/10.1016/j.tig.2016.01.003
Marquis-Nicholson R, Prosser DO, Love JM, Zhang L, Hayes I, George AM, Crawford JR, Skinner JR, Love DR (2014) Array comparative genomic hybridization identifies a heterozygous deletion of the entire KCNJ2 gene as a cause of sudden cardiac death. Circ Cardiovasc Genet 7(1):17–22. https://doi.org/10.1161/CIRCGENETICS.113.000415
Melo US, Schöpflin R, Acuna-Hidalgo R, Mensah MA, Fischer-Zirnsak B, Holtgrewe M, Klever MK, Türkmen S, Heinrich V, Pluym ID, Matoso E, Bernardo de Sousa S, Louro P, Hülsemann W, Cohen M, Dufke A, Latos-Bieleńska A, Vingron M, Kalscheuer V, Quintero-Rivera F, Spielmann M, Mundlos S (2020) Hi-C identifies complex genomic rearrangements and TAD-shuffling in developmental diseases. Am J Hum Genet 106(6):872–884. https://doi.org/10.1016/j.ajhg.2020.04.016
Micale L, Augello B, Maffeo C, Selicorni A, Zucchetti F, Fusco C, De Nittis P, Pellico MT, Mandriani B, Fischetto R, Boccone L, Silengo M, Biamino E, Perria C, Sotgiu S, Serra G, Lapi E, Neri M, Ferlini A, Cavaliere ML, Chiurazzi P, Monica MD, Scarano G, Faravelli F, Ferrari P, Mazzanti L, Pilotta A, Patricelli MG, Bedeschi MF, Benedicenti F, Prontera P, Toschi B, Salviati L, Melis D, Di Battista E, Vancini A, Garavelli L, Zelante L, Merla G (2014) Molecular analysis, pathogenic mechanisms, and readthrough therapy on a large cohort of Kabuki syndrome patients. Hum Mutat 35(7):841–850. https://doi.org/10.1002/humu.22547
Morlino S, Carbone A, Ritelli M, Fusco C, Giambra V, Nardella G, Notarangelo A, Panelli P, Mazzoccoli G, Zoppi N, Grammatico P, Wade EM, Colombi M, Castori M, Micale L (2019) TAB2 c.1398dup variant leads to haploinsufficiency and impairs extracellular matrix homeostasis. Hum Mutat 40(10):1886–1898. https://doi.org/10.1002/humu.23834
Mundlos S, Horn D (2014) Cooks syndrome. In: Limb malformations. Springer, Berlin, p 101
Palumbo O, Fichera M, Palumbo P, Rizzo R, Mazzolla E, Cocuzza DM, Carella M, Mattina T (2014) TBR1 is the candidate gene for intellectual disability in patients with a 2q24.2 interstitial deletion. Am J Med Genet A 164A(3):828–833. https://doi.org/10.1002/ajmg.a.36363
Palumbo O, Palumbo P, Di Muro E, Cinque L, Petracca A, Carella M, Castori M (2020) A private 16q24.2q24.3 microduplication in a boy with intellectual disability, speech delay and mild dysmorphic features. Genes (basel) 11(6):707. https://doi.org/10.3390/genes11060707
Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159(7):1665–1680. https://doi.org/10.1016/j.cell.2014.11.021
Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, Raca G, Ritter DI, South ST, Thorland EC, Pineda-Alvarez D, Aradhya S, Martin CL (2020) Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med 22(2):245–257. https://doi.org/10.1038/s41436-019-0686-8
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386. https://doi.org/10.1385/1-59259-192-2:365
Silva M, de Leeuw N, Mann K, Schuring-Blom H, Morgan S, Giardino D, Rack K, Hastings R (2019) European guidelines for constitutional cytogenomic analysis. Eur J Hum Genet 27:1–16. https://doi.org/10.1038/s41431-018-0244-x
Spielmann M, Mundlos S (2013) Structural variations, the regulatory landscape of the genome and their alteration in human disease. BioEssays 35(6):533–543. https://doi.org/10.1002/bies.201200178
Acknowledgements
The authors thank the family for their kind availability in sharing the findings within the scientific community.
Funding
This study was funded by the Italian Ministry of Health Ricerca Corrente 2018–2021.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest for this paper.
Ethics approval
This study was approved by the local Ethics Committee (protocol no. 13/CE/2021).
Consent to participate
The patient provided written informed consent for participation in this study.
Consent for publication
The patient signed a written informed consent for publication of molecular, clinical data and photographs.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
439_2021_2403_MOESM1_ESM.pdf
Figure S1 results of SNP-Array analysis in the patient and her parents for the 1p32.1 locus. Copy number state and Log2ratio value of each probe is drawn along chromosome band 1p32.1 (UCSC Genome Browser, build GRCh38/hg38). The upper panel (green) represents the genomic profile of the proband, the middle panel (orange) that of the mother and the lower panel (blue) that of the father. Values of Y-axis indicate the inferred copy number according the probes intensities. Arrows indicate the 1p32.1 chromosomal region deleted in the patient, covered by 7 SNP array probes (C-4LHXN, C-7JZTB, S-4JLOO, S-4QMGC, S-3EEPD, C-5MLLL, C-5CRVZ) (PDF 24 KB)
439_2021_2403_MOESM2_ESM.pdf
Figure S2 qPCR analysis of endogenous KCNJ16, SLC39A11, FGGY, DOCK7, and TAB2 transcript levels in patients’ and controls’ lymphocytes and fibroblasts. The graphs show data from two independent experiments. Each point is averaged over three technical replicates (PDF 383 KB)
Rights and permissions
About this article
Cite this article
Cinque, L., Micale, L., Manara, E. et al. A novel complex genomic rearrangement affecting the KCNJ2 regulatory region causes a variant of Cooks syndrome. Hum Genet 141, 217–227 (2022). https://doi.org/10.1007/s00439-021-02403-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-021-02403-y