Abstract
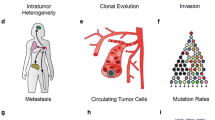
Intratumor heterogeneity within individual cancer tissues underlies the numerous phenotypes of cancer. Tumor subclones ultimately affect therapeutic outcomes due to their distinct molecular features. Drug-resistant subclones are present at a low frequency in tissues at the time of biopsy, but can also arise as a result of acquired somatic mutations. A number of different approaches have been utilized to understand the nature of intratumor heterogeneity. Clonal analysis using whole exome or genome sequencing data can help monitor subclones in the context of tumor progression. Multiregional biopsies permit the molecular characterization of subclones within tumors. Deep sequencing has also provided researchers with the ability to measure the low allele fraction variant within a small number of cells. Ultimately, single-cell sequencing will enable the identification of every minor population within a tumor microenvironment. In the clinical context, the ability to identify and monitor the subclonal architecture of a tumor is valuable for the development of precise cancer therapeutic methods.


Similar content being viewed by others
References
Alix-Panabieres C, Pantel K (2014) Challenges in circulating tumour cell research. Nat Rev Cancer 14:623–631. doi:10.1038/nrc3820
Andor N, Harness JV, Muller S, Mewes HW, Petritsch C (2014) EXPANDS: expanding ploidy and allele frequency on nested subpopulations. Bioinformatics 30:50–60. doi:10.1093/bioinformatics/btt622
Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, Ji HP, Maley CC (2016) Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 22:105–113. doi:10.1038/nm.3984
Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal SA, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Griffin CA, Burton J, Swerdlow H, Quail MA, Stratton MR, Iacobuzio-Donahue C, Futreal PA (2010) The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467:1109–1113. doi:10.1038/nature09460
Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, Beroukhim R, Pellman D, Levine DA, Lander ES, Meyerson M, Getz G (2012) Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 30:413–421. doi:10.1038/nbt.2203
Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G (2013) Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31:213–219. doi:10.1038/nbt.2514
de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, Jamal-Hanjani M, Shafi S, Murugaesu N, Rowan AJ, Gronroos E, Muhammad MA, Horswell S, Gerlinger M, Varela I, Jones D, Marshall J, Voet T, Van Loo P, Rassl DM, Rintoul RC, Janes SM, Lee SM, Forster M, Ahmad T, Lawrence D, Falzon M, Capitanio A, Harkins TT, Lee CC, Tom W, Teefe E, Chen SC, Begum S, Rabinowitz A, Phillimore B, Spencer-Dene B, Stamp G, Szallasi Z, Matthews N, Stewart A, Campbell P, Swanton C (2014) Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346:251–256. doi:10.1126/science.1253462
Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, McMichael JF, Wallis JW, Lu C, Shen D, Harris CC, Dooling DJ, Fulton RS, Fulton LL, Chen K, Schmidt H, Kalicki-Veizer J, Magrini VJ, Cook L, McGrath SD, Vickery TL, Wendl MC, Heath S, Watson MA, Link DC, Tomasson MH, Shannon WD, Payton JE, Kulkarni S, Westervelt P, Walter MJ, Graubert TA, Mardis ER, Wilson RK, DiPersio JF (2012) Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481:506–510. doi:10.1038/nature10738
Ding L, Wendl MC, McMichael JF, Raphael BJ (2014) Expanding the computational toolbox for mining cancer genomes. Nat Rev Genet 15:556–570. doi:10.1038/nrg3767
Falck AK, Ferno M, Bendahl PO, Ryden L (2013) St Gallen molecular subtypes in primary breast cancer and matched lymph node metastases–aspects on distribution and prognosis for patients with luminal A tumours: results from a prospective randomised trial. BMC Cancer 13:558. doi:10.1186/1471-2407-13-558
Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366:883–892. doi:10.1056/NEJMoa1113205
Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos CR, Martinez P, Phillimore B, Begum S, Rabinowitz A, Spencer-Dene B, Gulati S, Bates PA, Stamp G, Pickering L, Gore M, Nicol DL, Hazell S, Futreal PA, Stewart A, Swanton C (2014) Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 46:225–233. doi:10.1038/ng.2891
Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, Hicks DG, Lester S, Love R, Mangu PB, McShane L, Miller K, Osborne CK, Paik S, Perlmutter J, Rhodes A, Sasano H, Schwartz JN, Sweep FC, Taube S, Torlakovic EE, Valenstein P, Viale G, Visscher D, Wheeler T, Williams RB, Wittliff JL, Wolff AC (2010) American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol 28:2784–2795. doi:10.1200/JCO.2009.25.6529
Hiley C, de Bruin EC, McGranahan N, Swanton C (2014) Deciphering intratumor heterogeneity and temporal acquisition of driver events to refine precision medicine. Genome Biol 15:453. doi:10.1186/s13059-014-0453-8
Iwamoto T, Booser D, Valero V, Murray JL, Koenig K, Esteva FJ, Ueno NT, Zhang J, Shi W, Qi Y, Matsuoka J, Yang EJ, Hortobagyi GN, Hatzis C, Symmans WF, Pusztai L (2012) Estrogen receptor (ER) mRNA and ER-related gene expression in breast cancers that are 1 % to 10 % ER-positive by immunohistochemistry. J Clin Oncol 30:729–734. doi:10.1200/JCO.2011.36.2574
Kim KT, Lee HW, Lee HO, Kim SC, Seo YJ, Chung W, Eum HH, Nam DH, Kim J, Joo KM, Park WY (2015) Single-cell mRNA sequencing identifies subclonal heterogeneity in anti-cancer drug responses of lung adenocarcinoma cells. Genome Biol 16:127. doi:10.1186/s13059-015-0692-3
Kreso A, O’Brien CA, van Galen P, Gan OI, Notta F, Brown AM, Ng K, Ma J, Wienholds E, Dunant C, Pollett A, Gallinger S, McPherson J, Mullighan CG, Shibata D, Dick JE (2013) Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science 339:543–548. doi:10.1126/science.1227670
Kumar A, Boyle EA, Tokita M, Mikheev AM, Sanger MC, Girard E, Silber JR, Gonzalez-Cuyar LF, Hiatt JB, Adey A, Lee C, Kitzman JO, Born DE, Silbergeld DL, Olson JM, Rostomily RC, Shendure J (2014) Deep sequencing of multiple regions of glial tumors reveals spatial heterogeneity for mutations in clinically relevant genes. Genome Biol 15:530. doi:10.1186/s13059-014-0530-z
Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, Wan Y, Zhang W, Shukla SA, Vartanov A, Fernandes SM, Saksena G, Cibulskis K, Tesar B, Gabriel S, Hacohen N, Meyerson M, Lander ES, Neuberg D, Brown JR, Getz G, Wu CJ (2013) Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152:714–726. doi:10.1016/j.cell.2013.01.019
Lee MC, Lopez-Diaz FJ, Khan SY, Tariq MA, Dayn Y, Vaske CJ, Radenbaugh AJ, Kim HJ, Emerson BM, Pourmand N (2014) Single-cell analyses of transcriptional heterogeneity during drug tolerance transition in cancer cells by RNA sequencing. Proc Natl Acad Sci USA 111:E4726–E4735. doi:10.1073/pnas.1404656111
Leiserson MD, Vandin F, Wu HT, Dobson JR, Eldridge JV, Thomas JL, Papoutsaki A, Kim Y, Niu B, McLellan M, Lawrence MS, Gonzalez-Perez A, Tamborero D, Cheng Y, Ryslik GA, Lopez-Bigas N, Getz G, Ding L, Raphael BJ (2015) Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet 47:106–114. doi:10.1038/ng.3168
Li B, Li JZ (2014) A general framework for analyzing tumor subclonality using SNP array and DNA sequencing data. Genome Biol 15:473. doi:10.1186/s13059-014-0473-4
Miller CA, White BS, Dees ND, Griffith M, Welch JS, Griffith OL, Vij R, Tomasson MH, Graubert TA, Walter MJ, Ellis MJ, Schierding W, DiPersio JF, Ley TJ, Mardis ER, Wilson RK, Ding L (2014) SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput Biol 10:e1003665. doi:10.1371/journal.pcbi.1003665
Min JW, Kim WJ, Han JA, Jung YJ, Kim KT, Park WY, Lee HO, Choi SS (2015) Identification of distinct tumor subpopulations in lung adenocarcinoma via single-cell RNA-seq. PLoS One 10:e0135817. doi:10.1371/journal.pone.0135817
Navin N, Krasnitz A, Rodgers L, Cook K, Meth J, Kendall J, Riggs M, Eberling Y, Troge J, Grubor V, Levy D, Lundin P, Maner S, Zetterberg A, Hicks J, Wigler M (2010) Inferring tumor progression from genomic heterogeneity. Genome Res 20:68–80. doi:10.1101/gr.099622.109
Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, Muthuswamy L, Krasnitz A, McCombie WR, Hicks J, Wigler M (2011) Tumour evolution inferred by single-cell sequencing. Nature 472:90–94. doi:10.1038/nature09807
Ng CK, Martelotto LG, Gauthier A, Wen HC, Piscuoglio S, Lim RS, Cowell CF, Wilkerson PM, Wai P, Rodrigues DN, Arnould L, Geyer FC, Bromberg SE, Lacroix-Triki M, Penault-Llorca F, Giard S, Sastre-Garau X, Natrajan R, Norton L, Cottu PH, Weigelt B, Vincent-Salomon A, Reis-Filho JS (2015) Intra-tumor genetic heterogeneity and alternative driver genetic alterations in breast cancers with heterogeneous HER2 gene amplification. Genome Biol 16:107. doi:10.1186/s13059-015-0657-6
Oesper L, Mahmoody A, Raphael BJ (2013) THetA: inferring intra-tumor heterogeneity from high-throughput DNA sequencing data. Genome Biol 14:R80. doi:10.1186/gb-2013-14-7-r80
Oesper L, Satas G, Raphael BJ (2014) Quantifying tumor heterogeneity in whole-genome and whole-exome sequencing data. Bioinformatics 30:3532–3540. doi:10.1093/bioinformatics/btu651
Osborne CK, Schiff R (2011) Mechanisms of endocrine resistance in breast cancer. Annu Rev Med 62:233–247. doi:10.1146/annurev-med-070909-182917
Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suva ML, Regev A, Bernstein BE (2014) Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344:1396–1401. doi:10.1126/science.1254257
Qiao Y, Quinlan AR, Jazaeri AA, Verhaak RG, Wheeler DA, Marth GT (2014) SubcloneSeeker: a computational framework for reconstructing tumor clone structure for cancer variant interpretation and prioritization. Genome Biol 15:443. doi:10.1186/s13059-014-0443-x
Robasky K, Lewis NE, Church GM (2014) The role of replicates for error mitigation in next-generation sequencing. Nat Rev Genet 15:56–62. doi:10.1038/nrg3655
Roth A, Khattra J, Yap D, Wan A, Laks E, Biele J, Ha G, Aparicio S, Bouchard-Cote A, Shah SP (2014) PyClone: statistical inference of clonal population structure in cancer. Nat Methods 11:396–398. doi:10.1038/nmeth.2883
Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA (2012) Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci USA 109:14508–14513. doi:10.1073/pnas.1208715109
Sottoriva A, Kang H, Ma Z, Graham TA, Salomon MP, Zhao J, Marjoram P, Siegmund K, Press MF, Shibata D, Curtis C (2015) A Big Bang model of human colorectal tumor growth. Nat Genet 47:209–216. doi:10.1038/ng.3214
Spencer DH, Tyagi M, Vallania F, Bredemeyer AJ, Pfeifer JD, Mitra RD, Duncavage EJ (2014) Performance of common analysis methods for detecting low-frequency single nucleotide variants in targeted next-generation sequence data. J Mol Diagn 16:75–88. doi:10.1016/j.jmoldx.2013.09.003
Swanton C (2012) Intratumor heterogeneity: evolution through space and time. Cancer Res 72:4875–4882. doi:10.1158/0008-5472.CAN-12-2217
Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K, Larson DE, McLellan MD, Dooling D, Abbott R, Fulton R, Magrini V, Schmidt H, Kalicki-Veizer J, O’Laughlin M, Fan X, Grillot M, Witowski S, Heath S, Frater JL, Eades W, Tomasson M, Westervelt P, DiPersio JF, Link DC, Mardis ER, Ley TJ, Wilson RK, Graubert TA (2012) Clonal architecture of secondary acute myeloid leukemia. N Engl J Med 366:1090–1098. doi:10.1056/NEJMoa1106968
Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, Chen K, Scheet P, Vattathil S, Liang H, Multani A, Zhang H, Zhao R, Michor F, Meric-Bernstam F, Navin NE (2014) Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512:155–160. doi:10.1038/nature13600
Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N (2012) LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 40:11189–11201. doi:10.1093/nar/gks918
Wyatt AW, Mo F, Wang K, McConeghy B, Brahmbhatt S, Jong L, Mitchell DM, Johnston RL, Haegert A, Li E, Liew J, Yeung J, Shrestha R, Lapuk AV, McPherson A, Shukin R, Bell RH, Anderson S, Bishop J, Hurtado-Coll A, Xiao H, Chinnaiyan AM, Mehra R, Lin D, Wang Y, Fazli L, Gleave ME, Volik SV, Collins CC (2014) Heterogeneity in the inter-tumor transcriptome of high risk prostate cancer. Genome Biol 15:426. doi:10.1186/s13059-014-0426-y
Xu H, DiCarlo J, Satya RV, Peng Q, Wang Y (2014) Comparison of somatic mutation calling methods in amplicon and whole exome sequence data. BMC Genom 15:244. doi:10.1186/1471-2164-15-244
Yan W, Shih J, Rodriguez-Canales J, Tangrea MA, Player A, Diao L, Hu N, Goldstein AM, Wang J, Taylor PR, Lippman SM, Wistuba II, Emmert-Buck MR, Erickson HS (2013) Three-dimensional mRNA measurements reveal minimal regional heterogeneity in esophageal squamous cell carcinoma. Am J Pathol 182:529–539. doi:10.1016/j.ajpath.2012.10.028
Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C (2012) Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med 4:127ps10. doi:10.1126/scitranslmed.3003854
Yau C, Mouradov D, Jorissen RN, Colella S, Mirza G, Steers G, Harris A, Ragoussis J, Sieber O, Holmes CC (2010) A statistical approach for detecting genomic aberrations in heterogeneous tumor samples from single nucleotide polymorphism genotyping data. Genome Biol 11:R92. doi:10.1186/gb-2010-11-9-r92
Zhang J, Fujimoto J, Wedge DC, Song X, Seth S, Chow CW, Cao Y, Gumbs C, Gold KA, Kalhor N, Little L, Mahadeshwar H, Moran C, Protopopov A, Sun H, Tang J, Wu X, Ye Y, William WN, Lee JJ, Heymach JV, Hong WK, Swisher S, Wistuba II, Futreal PA (2014) Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 346:256–259. doi:10.1126/science.1256930
Acknowledgments
This work was supported by Health Technology R&D Project, the Ministry of Health and Welfare (HI13C2096).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Rights and permissions
About this article
Cite this article
Ryu, D., Joung, JG., Kim, N.K.D. et al. Deciphering intratumor heterogeneity using cancer genome analysis. Hum Genet 135, 635–642 (2016). https://doi.org/10.1007/s00439-016-1670-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-016-1670-x