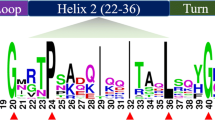
Abstract
Tea plant (Camellia sinensis L.), whose leaves are the major reproductive organs, has been cultivated and consumed widely for its economic and health benefits. The Knotted1-like Homeobox (KNOX) proteins play significant roles in leaf morphology formation and development. However, the functions of KNOX proteins in tea plants are still unknown. Here, 11 CsKNOX genes from the tea plants were cloned and divided into Class I, II, and KNATM clades based on their protein sequences. These 11 CsKNOX genes were mapped on 8 out of 15 tea plant chromosomes, all localized in the nucleus. Specific spatiotemporal expression patterns of CsKNOX genes were found in various tissues and different development periods of buds, flowers, and roots of tea plants. Meanwhile, transcript levels of CsKNOX in tea leaves were strongly correlated with the accumulation of flavan-3-ols and proanthocyanidins. It was found that most of the CsKNOX genes could respond to drought, salt, cold, and exogenous MeJA and GA3 by analysis of transcriptomics data and promoter elements. The protein interaction analysis showed that CsKNOX could cooperate with CsAS1 and other critical functional proteins. In conclusion, this research provided the basic information for the functions of the CsKNOX family during organogenesis and stress response in tea plants, which was necessary for further functional characterization verification.












Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article.
References
Bar M, Ori N (2014) Leaf development and morphogenesis. Development 141:4219–4230. https://doi.org/10.1242/dev.106195
Brody H (2019) Tea. Nature 566(7742):S1. https://doi.org/10.1038/d41586-019-00394-5
Champagne CEM, Goliber TE, Wojciechowski MF, Mei RW, Townsley BT, Wang K, Paz MM, Geeta R, Sinha NR (2007) Compound leaf development and evolution in the legumes. Plant Cell 19:3369–3378. https://doi.org/10.1105/tpc.107.052886
Chatterjee M, Bermudez-Lozano CL, Clancy M, Davis TM, Folta KM (2011) A strawberry KNOX gene regulates leaf, flower, and meristem architecture. PLoS ONE 6:e24752. https://doi.org/10.1371/journal.pone.0024752
Chen D, Molitor AM, Xu L, Shen WH (2016) Arabidopsis PRC1 core component AtRING1 regulates stem cell-determining carpel development mainly through repression of class I KNOX genes. BMC Biol 14:112. https://doi.org/10.1186/s12915-016-0336-4
Cheng X, Li M, Abdullah M, Li G, Zhang J, Manzoor MA, Wang H, Jin Q, Jiang T, Cai Y, Li D, Lin Y (2019) In silico genome-wide analysis of the pear (Pyrus bretschneideri) KNOX family and the functional characterization of PbKNOX1, an Arabidopsis BREVIPEDICELLUS orthologue gene, involved in cell wall and lignin biosynthesis. Front Genet 10:632. https://doi.org/10.3389/fgene.2019.00632
Chuck G, Lincoln C, Hake S (1996) KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8:1277–1289. https://doi.org/10.1105/tpc.8.8.1277
Ezura K, Nakamura A, Mitsuda N (2022) Genome-wide characterization of the TALE homeodomain family and the KNOX-BLH interaction network in tomato. Plant Mol Biol 109:799–821. https://doi.org/10.1007/s11103-022-01277-6
Fleming AJ (2002) The mechanism of leaf morphogenesis. Planta 216:17–22. https://doi.org/10.1007/s00425-002-0864-8
Gao J, Yang X, Zhao W, Lang T, Samuelsson T (2015) Evolution, diversification, and expression of KNOX proteins in plants. Front Plant Sci 6:882. https://doi.org/10.3389/fpls.2015.00882
Guo K, Huang C, Miao Y, Cosgrove DJ, Hsia KJ (2022) Leaf morphogenesis: the multifaceted roles of mechanics. Mol Plant 15:1098–1119. https://doi.org/10.1016/j.molp.2022.05.015
Gupta S, Rosenthal DM, Stinchcombe JR, Baucom RS (2020) The remarkable morphological diversity of leaf shape in sweet potato (Ipomoea batatas): the influence of genetics, environment, and GxE. New Phytol 225:2183–2195. https://doi.org/10.1111/nph.16286
Hake S, Ori N (2002) Plant morphogenesis and KNOX genes. Nat Genet 31:121–122. https://doi.org/10.1038/ng0602-121
Hake S, Char BR, Chuck G, Foster T, Long J, Jackson D (1995) Homeobox genes in the functioning of plant meristems. Philos Trans R Soc Lond B Biol Sci 350:45–51. https://doi.org/10.1098/rstb.1995.0136
Hake S, Smith HMS, Holtan H, Magnani E, Mele G, Ramirez J (2004) The role of KNOX genes in plant development. Annu Rev Cell Dev Biol 20:125–151. https://doi.org/10.1146/annurev.cellbio.20.031803.093824
Hasson A, Blein T, Laufs P (2010) Leaving the meristem behind: the genetic and molecular control of leaf patterning and morphogenesis. CR Biol 333:350–360. https://doi.org/10.1016/j.crvi.2010.01.013
Hay A, Tsiantis M (2006) The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat Genet 38:942–947. https://doi.org/10.1038/ng1835
Hay A, Tsiantis M (2009) A KNOX family TALE. Curr Opin Plant Biol 12:593–598. https://doi.org/10.1016/j.pbi.2009.06.006
Hay A, Tsiantis M (2010) KNOX genes: versatile regulators of plant development and diversity. Development 137:3153–3165. https://doi.org/10.1242/dev.030049
Ikezaki M, Kojima M, Sakakibara H, Kojima S, Ueno Y, Machida C, Machida Y (2010) Genetic networks regulated by ASYMMETRIC LEAVES1 (AS1) and AS2 in leaf development in Arabidopsis thaliana: KNOX genes control five morphological events. Plant J 61:70–82. https://doi.org/10.1111/j.1365-313X.2009.04033.x
Izhaki A, Bowman JL (2007) KANADI and class III HD-zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19:495–508. https://doi.org/10.1105/tpc.106.047472
Jasinski S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M (2005) KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol 15:1560–1565. https://doi.org/10.1016/j.cub.2005.07.023
Jia P, Zhang CG, Xing LB, Li YM, Shah KR, Zuo XY, Zhang D, An N, Han MY, Ren XL (2020) Genome-wide identification of the MdKNOX gene family and characterization of its transcriptional regulation in Malus domestica. Front Plant Sci 11:128. https://doi.org/10.3389/fpls.2020.00128
Jia P, Xing L, Zhang C, Chen H, Li Y, Zhang D, Ma J, Zhao C, Han M, Ren X, An N (2021) MdKNOX15, a class I knotted-like transcription factor of apple, controls flowering and plant height by regulating GA levels through promoting the MdGA2ox7 transcription. Environ Exp Bot 185:104411. https://doi.org/10.1016/j.envexpbot.2021.104411
Kerstetter R, Vollbrecht E, Lowe B, Veit B, Yamaguchi J, Hake S (1994) Sequence-analysis and expression patterns divide the maize knotted1-like homeobox genes into 2 classes. Plant Cell 6:1877–1887. https://doi.org/10.1105/tpc.6.12.1877
Koltai H, Bird DM (2000) Epistatic repression of PHANTASTICA and class 1 KNOTTED genes is uncoupled in tomato. Plant J 22:455–459. https://doi.org/10.1046/j.1365-313X.2000.00754.x
Lebedeva OV, Ezhova TA, Melzer S (2005) Gene TAENIATA is a novel negative regulator of the Arabidopsis thaliana homeobox genes KNAT1, KNAT2, KNAT6, and STM. Russ J Genet 41:871–876. https://doi.org/10.1007/s11177-005-0174-x
Li LC, Kang DM, Chen ZL, Qu LJ (2007) Hormonal regulation of leaf morphogenesis in Arabidopsis. J Integr Plant Biol 49:75–80. https://doi.org/10.1111/j.1744-7909.2006.00410.x
Li EY, Wang SC, Liu YY, Chen JG, Douglas CJ (2011) OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J 67:328–341. https://doi.org/10.1111/j.1365-313X.2011.04595.x
Liu J, Ha D, Xie Z, Wang C, Wang H, Zhang W, Zhang J, Chen S (2008) Ectopic expression of soybean GmKNT1 in Arabidopsis results in altered leaf morphology and flower identity. J Genet Genomics 35:441–449
Liu J, Zhang C, Han J, Fang X, Xu H, Liang C, Li D, Yang Y, Cui Z, Wang R, Song J (2022) Genome-wide analysis of KNOX transcription factors and expression pattern of dwarf-related KNOX genes in pear. Front Plant Sci. https://doi.org/10.3389/fpls.2022.806765
Mantegazza R, Tononi P, Moller M, Spada A (2009) WUS and STM homologs are linked to the expression of lateral dominance in the acaulescent Streptocarpus rexii (Gesneriaceae). Planta 230:529–542. https://doi.org/10.1007/s00425-009-0965-8
Meisel L, Lam E (1996) The conserved ELK-homeodomain of KNOTTED-1 contains two regions that signal nuclear localization. Plant Mol Biol 30:1–14. https://doi.org/10.1007/BF00017799
Meng L, Liu X, He C, Xu B, Li Y, Hu Y (2020) Functional divergence and adaptive selection of KNOX gene family in plants. Open Life Sci 15:346–363. https://doi.org/10.1515/biol-2020-0036
Nagasaki H, Sakamoto T, Sato Y, Matsuoka M (2001) Functional analysis of the conserved domains of a rice KNOX homeodomain protein, OSH15. Plant Cell 13:2085–2098. https://doi.org/10.1105/TPC.010113
Nishii K, Moeller M, Kidner C, Spada A, Mantegazza R, Wang CN, Nagata T (2010) A complex case of simple leaves: indeterminate leaves co-express ARP and KNOX1 genes. Dev Genes Evol 220:25–40. https://doi.org/10.1007/s00427-010-0326-4
Ortiz-Ramírez CI, Plata-Arboleda S, Pabón-Mora N (2018) Evolution of genes associated with gynoecium patterning and fruit development in Solanaceae. Ann Bot 121:1211–1230. https://doi.org/10.1093/aob/mcy007
Pagnussat GC, Yu HJ, Sundaresana V (2007) Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 19:3578–3592. https://doi.org/10.1105/tpc.107.054890
Pan SY, Nie Q, Tai HC, Song XL, Tong YF, Zhang LJF, Wu XW, Lin ZH, Zhang YY, Ye DY, Zhang Y, Wang XY, Zhu PL, Chu ZS, Yu ZL, Liang C (2022) Tea and tea drinking: China’s outstanding contributions to the mankind. Chin Med 17:1–40. https://doi.org/10.1186/s13020-022-00571-1
Pekker I, Alvarez JP, Eshed Y (2005) Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17:2899–2910. https://doi.org/10.1105/tpc.105.034876
Rast MI, Simon R (2012) Arabidopsis JAGGED LATERAL ORGANS acts with ASYMMETRIC LEAVES2 to coordinate KNOX and PIN expression in shoot and root meristems. Plant Cell 24:2917–2933. https://doi.org/10.1105/tpc.112.099978
Rathour M, Alok A, Upadhyay SK (2022) Investigation of roles of TaTALE genes during development and stress response in bread wheat. Plants Basel 11:587. https://doi.org/10.3390/plants11050587
Rutjens B, Bao DP, Van Eck-Stouten E, Brand M, Smeekens S, Proveniers M (2009) Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J 58:641–654. https://doi.org/10.1111/j.1365-313X.2009.03809.x
Sahaf M, Sharon E (2016) The rheology of a growing leaf: stress-induced changes in the mechanical properties of leaves. J Exp Bot 67:5509–5515. https://doi.org/10.1093/jxb/erw316
Saidi A, Hajibarat Z (2021) Genome wide identification of StKNOX gene family and characterization of their expression in Solanum tuberosum. Biocatal Agric Biotechnol 37:102160. https://doi.org/10.1016/j.bcab.2021.102160
Sakamoto T, Nishimura A, Tamaoki M, Kuba M, Tanaka H, Iwahori S, Matsuoka M (1999) The conserved KNOX domain mediates specificity of tobacco KNOTTED1-type homeodomain proteins. Plant Cell 11:1419–1431. https://doi.org/10.1105/tpc.11.8.1419
Scanlon MJ, Henderson DC, Bernstein B (2002) SEMAPHORE1 functions during the regulation of ancestrally duplicated KNOX genes and polar auxin transport in maize. Development 129:2663–2673. https://doi.org/10.1242/dev.129.11.2663
Scofield S, Murray JAH (2006) KNOX gene function in plant stem cell niches. Plant Mol Biol 60:929–946. https://doi.org/10.1007/s11103-005-4478-y
Scofield S, Dewitte W, Murray JAH (2008) A model for Arabidopsis class-1 KNOX gene function. Plant Signal Behav 3:257–259. https://doi.org/10.4161/psb.3.4.5194
Scofield S, Dewitte W, Murray JAH (2014) STM sustains stem cell function in the Arabidopsis shoot apical meristem and controls KNOX gene expression independently of the transcriptional repressor AS1. Plant Signal Behav 9:e28934. https://doi.org/10.4161/psb.28934
Shani E, Yanai O, Ori N (2006) The role of hormones in shoot apical meristem function. Curr Opin Plant Biol 9:484–489. https://doi.org/10.1016/j.pbi.2006.07.008
Shi J, Ma CY, Qi DD, Lv HP, Yang T, Peng QH, Chen ZM, Lin Z (2015) Transcriptional responses and flavor volatiles biosynthesis in methyl jasmonate-treated tea leaves. BMC Plant Biol 15:233. https://doi.org/10.1186/s12870-015-0609-z
Smith HMS, Hake S (2003) The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15:1717–1727. https://doi.org/10.1105/tpc.012856
Song X, Zhao Y, Wang J, Lu MZ (2021) The transcription factor KNAT2/6b mediates changes in plant architecture in response to drought via down-regulating GA20ox1 in Populus alba x P. glandulosa. J Exp Bot 72:5625–5637. https://doi.org/10.1093/jxb/erab201
Srinivasan C, Liu Z, Scorza R (2011) Ectopic expression of class 1 KNOX genes induces adventitious shoot regeneration and alter growth and development of tobacco (Nicotiana tabacum L) and European plum (Prunus domestica L). Plant Cell Rep 30:655–664. https://doi.org/10.1007/s00299-010-0993-7
Tai YL, Liu C, Yu SW, Yang H, Sun JM, Guo CX, Huang B, Liu ZY, Yuan Y, Xia EH, Wei CL, Wan XC (2018) Gene co-expression network analysis reveals coordinated regulation of three characteristic secondary biosynthetic pathways in tea plant (Camellia sinensis). BMC Genomics 19:1–13. https://doi.org/10.1186/s12864-018-4999-9
Tanaka M, Kato N, Nakayama H, Nakatani M, Takahata Y (2008) Expression of class I knotted1-like homeobox genes in the storage roots of sweetpotato (Ipomoea batatas). J Plant Physiol 165:1726–1735. https://doi.org/10.1016/j.jplph.2007.11.009
Testone G, Condello E, Verde I, Nicolodi C, Caboni E, Dettori MT, Vendramin E, Bruno L, Bitonti MB, Mele G, Giannino D (2012) The peach (Prunus persica L. Batsch) genome harbours 10 KNOX genes, which are differentially expressed in stem development, and the class 1 KNOPE1 regulates elongation and lignification during primary growth. J Exp Bot 63:5417–5435. https://doi.org/10.1093/jxb/ers194
Timmermans MCP, Hudson A, Becraft PW, Nelson T (1999) ROUGH SHEATH2: a Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284:151–153
Uchida N, Kimura S, Koenig D, Sinha N (2010) Coordination of leaf development via regulation of KNOX1 genes. J Plant Res 123:7–14. https://doi.org/10.1007/s10265-009-0248-2
Wang Y, Jiao Y (2020) Keeping leaves in shape. Nat Plants 6:436–437. https://doi.org/10.1038/s41477-020-0660-0
Wang XC, Zhao QY, Ma CL, Zhang ZH, Cao HL, Kong YM, Yue C, Hao XY, Chen L, Ma JQ, Jin JQ, Li X, Yang YJ (2013) Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genomics 14:1–15. https://doi.org/10.1186/1471-2164-14-415
Wei CL, Yang H, Wang SB et al (2018) Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc Nat Acad Sci 115(18): E4151–E4158
Woerlen N, Allam G, Popescu A, Corrigan L, Pautot V, Hepworth SR (2017) Repression of BLADE-ON-PETIOLE genes by KNOX homeodomain protein BREVIPEDICELLUS is essential for differentiation of secondary xylem in Arabidopsis root. Planta 245:1079–1090. https://doi.org/10.1007/s00425-017-2663-2
Xi O, Yudan W, Jingyi L, Jiewen Zh, Zhenbin X, Bing H, Zhehao J, Yuting W, Wei S, Shiwei S, Yanwei H, Riyuan Ch (2022) Genome-wide identification of the KNOTTED HOMEOBOX gene family and their involvement in stalk development in flowering Chinese cabbage. Front Plant Sci 72:7002–7019. https://doi.org/10.3389/fpls.2022.1019884
Yang JY, Iwasaki M, Machida C, Machida Y, Zhou X, Chua NH (2008) beta C1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev 22:2564–2577. https://doi.org/10.1101/gad.1682208
Zeng RF, Zhou H, Fu LM, Yan Z, Ye LX, Hu SF, Gan ZM, Ai XY, Hu CG, Zhang JZ (2021) Two citrus KNAT-like genes, CsKN1 and CsKN2, are involved in the regulation of spring shoot development in sweet orange. J Exp Bot 72(20):7002–7019. https://doi.org/10.1093/jxb/erab311
Zhang Q, Cai MC, Yu XM, Wang LS, Guo CF, Ming R, Zhang JS (2017) Transcriptome dynamics of Camellia sinensis in response to continuous salinity and drought stress. Tree Genet Genomes 13(4):1–17. https://doi.org/10.1007/s11295-017-1161-9
Zhang SY, Pan YP, Zhi CC, Zheng YJ, Wang XA, Li XX, Cheng ZH (2021a) Genome-wide identification and characterization of KNOTTED-like homeobox (KNOX) homologs in garlic (Allium sativum L.) and their expression profilings responding to exogenous cytokinin and gibberellin. Int J Mol Sci 22:9237. https://doi.org/10.3390/ijms22179237
Zhang X, Zhao J, Wu X, Hu G, Fan S, Ma Q (2021b) Evolutionary relationships and divergence of KNOTTED1-like family genes involved in salt tolerance and development in cotton (Gossypium hirsutum L.). Front Plant Sci 12:774161. https://doi.org/10.3389/fpls.2021.774161
Zhang Y, Yin Q, Qin W, Gao H, Du J, Chen J, Li H, Zhou G, Wu H, Wu AM (2022) The Class II KNOX family members KNAT3 and KNAT7 redundantly participate in Arabidopsis seed coat mucilage biosynthesis. J Exp Bot 73:3477–3495. https://doi.org/10.1093/jxb/erac066
Zhou R, Fan M, Zhao M, Jiang X, Liu Q (2022) Overexpression of LtKNOX1 from Lilium tsingtauense in Nicotiana benthamiana affects the development of leaf morphology. Plant Signal Behav. https://doi.org/10.1080/15592324.2022.2031783
Acknowledgements
All the authors thank the public databases (TPIA) for the raw data downloaded and we thank all our colleagues in the laboratory for providing useful discussions and technical assistance. We are very grateful to Prof. Yongsheng Yu for critically evaluating the manuscript and providing constructive comments for its improvement.
Funding
This work was supported by the Chongqing Technology Innovation and Application Demonstration Project (cstc2021jscx-gksbX0016 and 2022TIAD-CUX0006) and the National Natural Science Foundation of China (31400583).
Author information
Authors and Affiliations
Contributions
LY and HT designed and supervised the project and reviewed drafts of the paper; HD conceived this study, performed all experiments, analyzed the data, and wrote the original draft manuscript; CZ provided experiment materials for expression analysis; SZ and RH collected the samples, and performed part of the experiments. LY provided suggestions for improvement of the experiments and the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Communicated by Bing Yang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dai, H., Zheng, S., Zhang, C. et al. Identification and expression analysis of the KNOX genes during organogenesis and stress responseness in Camellia sinensis (L.) O. Kuntze. Mol Genet Genomics 298, 1559–1578 (2023). https://doi.org/10.1007/s00438-023-02075-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-023-02075-5