Abstract
To better understand the interaction between SARS-CoV-2 and human host and find potential ways to block the pandemic, one of the unresolved questions is that how the virus economically utilizes the resources of the hosts. Particularly, the tRNA pool has been adapted to the host genes. If the virus intends to translate its own RNA, then it has to compete with the abundant host mRNAs for the tRNA molecules. Translation initiation is the rate-limiting step during protein synthesis. The tRNAs carrying the initiation Methionine (iMet) recognize the start codon termed initiation ATG (iATG). Other normal Met-carrying tRNAs recognize the internal ATGs. The tAI of virus genes is significantly lower than the tAI of human genes. This disadvantage in translation elongation of viral RNAs must be compensated by more efficient initiation rates. In the human genome, the abundance of iMet–tRNAs to Met–tRNAs is five times higher than the iATG to ATG ratio. However, when SARS-CoV-2 infects human cells, the iMet has an 8.5-time enrichment to iATG. We collected 58 virus species and found that the enrichment of iMet is higher in all viruses compared to human. Our study indicates that the genome sequences of viruses like SARS-CoV-2 have the advantage of competing for the iMet–tRNAs with host mRNAs. The capture of iMet–tRNAs allows the fast translation initiation and the reproduction of virus itself, which compensates the lower tAI of viral genes. This might explain why the virus could rapidly translate its own RNA and reproduce itself from the sea of host mRNAs. Meanwhile, our study reminds the researchers not to ignore the mutations related to ATGs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Understanding the interaction between virus and human host is important for finding potential approaches to fight against the virus. Among the various ways of host–parasite interactions, one of the unresolved issues is that how the virus economically utilizes the resources of the hosts.
In the beginning of year 2020, the outbreak of SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) has caused severe damage to China especially the Hubei province (Cowling and Leung 2020; Hui et al. 2020; Wang et al. 2020). Recently, the whole world is suffering from this pandemic. There is urgent need to understand the relationship and interaction between SARS-CoV-2 and the human hosts. It is plausible to study both the virus and human genomes in the light of evolution and adaptation.
It is well acknowledged that the RNA viruses translate its RNAs and reproduce their own proteins by using the resources from host cells. An unresolved question is how the virus economically utilizes the materials of the hosts? Take the translation process for instance. According to the early-established tRNA adaptation theory (dos Reis et al. 2004; Ikemura and Ozeki 1983), the tRNA pool of the host has been adapted to the codon usage of the host genes. If the RNA virus tries to translate its own RNA to produce its proteins, then it has to compete with the abundant host mRNAs for the tRNA resources.
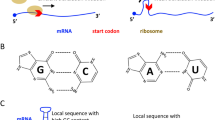
The efficiency of translation initiation and elongation determine the rate of protein synthesis. Particularly, initiation is the major rate-limiting step. In the coding sequence (CDS) of mRNAs, ATG appears in most of the start codon position but also appears in the body of CDS (Fig. 1a). We denote the initiation ATG as “iATG,” and other internal ATGs as “ATG.” The tRNAs carrying the initiation Methionine (iMet) recognize the iATG. Other normal Met-carrying tRNAs recognize the internal ATGs (Fig. 1a). When decoding a codon, the cognate tRNAs (the matched tRNAs) and other non-cognate tRNAs (the unmatched tRNAs) compete for base-pairing with the codon. The non-cognate tRNAs would finally be rejected by the ribosomes (Thompson et al. 1981; Thompson and Stone 1977). Thus, the ratio of cognate to non-cognate tRNAs is essential for the efficient translation of a codon. On the other hand, when a tRNA molecule is searching for its pairing codon, the ratio of matched codon to unmatched codon is important for the efficient searching and decoding.
Relationship between codons and tRNAs. a A diagram that illustrates the translation process involving iMet–tRNAs and Met–tRNAs. In the diagram, iMet–tRNA represents the tRNA-carrying initiation methionine. Met–tRNA is the tRNA-carrying normal internal Met. iATG represents the start codon (in most genes the start codon is ATG). ATG represents the internal ATG codon in the body of CDS. Other non-ATG codons in the CDS are omitted. b Pearson correlation between codon frequency in the human genome and the human tAI. c Pearson correlation between codon frequency in the SARS-CoV-2 genome and the human tAI
As we have mentioned above, the tRNA pool of host cells has been adapted to the codon usage of the hosts rather than the parasites. The codon composition of the virus genome is not optimal for translation in host cells at all. This fact could be reflected by the tRNA adaptation index (tAI) which measures the overall tRNA availability of a gene (dos Reis et al. 2004). So that the translation elongation of virus RNAs would suffer from low tRNA abundance and low efficiency. This does not mean that the protein synthesis process is blocked. Even the ribosomes elongate slowly on the viral RNA, one solution to enhance the global translation efficiency is to let more viral RNAs be translated simultaneously. In other words, the way to compensate the elongation deficiency is to enhance the initiation rate. For the translation initiation, the only way to ensure this process is to let iMet–tRNA find iATG rapidly and accurately. Other normal Met–tRNAs would compete for iATGs with iMet–tRNAs. Therefore, excessive non-initiation Met–tRNAs are not suitable for fast translation initiation.
We define the “enrichment of iMet” = (number of iMet/number of Met + iMet)/(number of iATG/number of internal ATGs + iATGs). Higher enrichment of iMet would facilitate the translation initiation and guarantee the fast production. If the RNA virus intends to survive and propagate itself in the host environment, then it has to compete for iMet–tRNAs with host cells. We first compared the enrichment of iMet in human and SARS-CoV-2. We found that this value is 5.2 and 8.5 in human and SARS-CoV-2, respectively. We also collected up to 58 virus species and constantly found that the enrichment of iMet is higher in all viruses compared to human. This might be a consequence of selection and evolution that ensures the virus to survive and propagate in the host systems.
Our current results suggest that the genomes of SARS-CoV-2 and other viruses have the advantage of competing for the iMet–tRNAs with host mRNAs. The capture of iMet–tRNAs by start codons allows the efficient translation initiation and fast reproduction of viral RNA and proteins. The higher efficiency in translation initiation would compensate the lower tAI of viral genes. Our study raised a possibility of how the virus could successfully survive in the host environment, translate its own RNA, and reproduce itself from the sea of host mRNAs.
Materials and methods
Data collection
We downloaded the novel coronavirus SARS-CoV-2 genome as well as other virus genomes from the NCBI website (https://www.ncbi.nlm.nih.gov/genome/). The coding sequences were extracted according to the genome annotation. The coding sequence of human genome was downloaded from the Ensembl website of version hg19 (ftp://ftp.ensembl.org/pub/release-75/fasta/homo_sapiens/cds/). The tRNA copy number in human genome is downloaded from the Genomic tRNA Database (https://gtrnadb.ucsc.edu/). The tRNA copy numbers have been used to roughly represent the tRNA abundance in plenty of early studies (dos Reis et al. 2004; Sabi and Tuller 2014).
Enrichment of iMet
We define the enrichment of iMet = (number of iMet/number of Met + iMet)/ (number of iATG/number of internal ATGs + iATGs).
Calculation of tAI
The calculation of tRNA adaptation index (tAI) (dos Reis et al. 2004) considered both the tRNA copy number and the wobble interaction between codon and anticodon. The weighted sum of tRNA copy number was assigned to each codon and then normalized by the maximum number among all codons. Thus, each codon has a copy number value which is normalized to 0 ~ 1. The tAI of a gene is the geometric mean of this value of each codon so that the final tAI of a gene also ranges from 0 to 1. Higher tAI value of a gene represents higher tRNA availability and higher translatability.
Statistical analyses
R language was used to perform the statistical analyses and graphic work.
Data availability
All data used in our study are public data.
SARS-CoV-2 genome and other virus’ genomes: NCBI website (https://www.ncbi.nlm.nih.gov/genome/).
The coding sequence of human genome: Ensembl website version hg19 (ftp://ftp.ensembl.org/pub/release-75/fasta/homo_sapiens/cds/).
The tRNA copy number in human genome: Genomic tRNA Database (https://gtrnadb.ucsc.edu/).
Results
tAI profile of human and viral genes
We downloaded the coding sequences of 58 virus species (including SARS-CoV-2) and human, and also obtained the tRNA species and copy numbers in human genome (“Materials and methods”). Each virus species has 5 ~ 14 coding genes according to the genome annotation. In human, there are twenty thousand unique coding genes. We calculated the tAI value of each human and viral genes (“Materials and methods”). First, as we have mentioned in the background, the tRNAs and codons adapt to each other to allow efficient decoding during translation elongation. We verified that in the human genome the codon usage and the corresponding tRNA copies are highly correlated (Fig. 1b). In contrast, codon usage in the virus genome does not correlate with human tRNA at all (Fig. 1c). Moreover, the tAI measurement could describe the tRNA availability at both codon level and gene level (Chu and Wei 2020; dos Reis et al. 2004; Sabi and Tuller 2014). From the distribution profile of gene level tAI, we could see that the viral genes have significantly lower tAI values than human genes (Fig. 2a).Using KS tests to determine the statistical significance, all of the 58 viruses have globally lower tAI values compared to human even after multiple testing correction (Benjamini and Hochberg 1995). It indicates that the codon usage of viral genes is not adapted to the tRNA pool of human host. This might not be surprising because the GC content of human genes is remarkably higher than the GC content in viruses, and therefore the optimal codons in humans may not appear so frequently in viral genes. As a result, the translation elongation process of viral RNAs is impeded. One way to compensate the deficiency in translation elongation is to increase the initiation rate. In other words, let more viral RNAs be translated simultaneously.
The tAI profile and the statistic of iMet–tRNAs and iATGs. a tAI values of genes in human and 58 virus species. The mean and standard error is plotted as circles and bars. The p values are calculated by KS tests. Asterisk “*” represents p value < 0.05 after multiple testing correction. b The statistic in human genome. The formula calculating the enrichment of iMet is provided in the Methods, and could also be understood from the figure
Parsing the Met–tRNAs and ATGs in the human genome
The definition of iMet, Met, iATG, and ATG has already been introduced in the Background (Fig. 1a). We use the tRNA copy numbers to represent the relative amount of tRNAs. Among the human tRNA copies annotated in the genome, 9 were annotated to carry iMet and 11 were annotated to carry internal Met. The proportion of iMet–tRNA is 45% (Fig. 2b). There are 20.8 thousand unique coding genes in the human genome so that there are 20.8 K iATGs. When we retrieved the longest CDS of each gene, there are totally 242.5 thousand internal ATGs. The proportion of iATGs is 8.6% (Fig. 2b). This result demonstrates that the proportion of iMet–tRNAs is 5.2 times higher than the proportion of iATGs in the human genome.
It is known that the copy number of tRNAs species is highly correlated with the amino acid and codon usage. However, iMet seems to be an exception. The abundance of iMet–tRNAs is always excessive compared to the relative amount of iATGs. By using Chi-square test on the number of iMet over iATG versus other AA-tRNA over other codons, we obtain a p value of 2e-10, which is very significant. This might reflect the urgent need for efficient translation initiation. Note that the classification of initiation and internal AA-tRNAs is only for ATG but not for other codons or amino acids due to its special function in translation initiation.
The enrichment of iMet in SARS-CoV-2 and other viruses
The viruses infect the host cells and utilize the resources of the host. The iMet–tRNA and Met–tRNA should be the same as the human host (Fig. 3a). The SARS-CoV-2 has 12 coding genes, among which ORF1a is completely included in the sequence of ORF1ab. The 11 non-redundant genes have 11 iATGs and 196 internal ATGs. The proportion of iATG is 5.3% and the enrichment of iMet is 8.5 (Fig. 3a).This enrichment value is considerably higher than the 5.2 in human.
The statistic of iMet–tRNAs and iATGs in SARS-CoV-2 and other viruses. a Statistics in SARS-CoV-2. The meaning of iMet, Met, iATG, and ATG has been described in Fig. 1 and the “Materials and methods” section. b Enrichment of iMet in 58 virus species (orange) and human (purple). The p values of enrichment in virus versus human are calculated with Fisher’s exact tests. FDR (false discovery rate) is the p value corrected by the multiple testing correction
We wonder whether the higher enrichment of iMet in SARS-CoV-2 is obtained by chance or it is a general trend for viruses. We downloaded the sequences of 58 different viruses (“Materials and methods”) and calculated the same parameter. Amazingly, the enrichment values of iMet are constantly higher than human (Fig. 3b). Using Fisher’s exact tests, we discovered 45 viruses with significantly higher enrichment of iMet compared to human, and 23 after multiple testing correction (Benjamini and Hochberg 1995) (Fig. 3b). These results demonstrate that the viruses naturally have an advantage of competing for the iMet–tRNAs required for translation initiation. This advantage perfectly compensates the deficiency in translation elongation caused by low tAI of viral genes.
Discussion
The SARS-CoV-2 needs to utilize the resources from host cells to reproduce itself. The abundance of tRNAs usually acts as the major limitation of translation. This is why the tRNA pool of an organism is correlated with its genomic codon usage rather than the codon usage in other species. Not surprisingly, the codon usage of virus is largely different from that of the host cells. This discrepancy between human host and viruses could be clearly seen from the profile of tAI values of genes. When the viral RNA is being translated in the host cells, the codons frequently used by the virus might not be optimal codons in the hosts. Therefore, the translation elongation of viral RNAs would suffer from scarce tRNAs and low decoding rates. The way to compensate the inefficient elongation is to let more viral RNAs be translated simultaneously. A higher initiation rate would accomplish this goal.
We have found that the virus sequences have intuitively higher enrichment of iMet to compete with the host mRNAs and allow for fast translation. Our idea is not restricted to the human–virus relationship. The enrichment of iMet could also be applied to other host–parasite systems. Hopefully, this hypothesis could be tested in a much larger range of species. It remains to be seen whether the parasite species always have higher enrichment of iMet compared to the host species. Moreover, in theory the mutations that increase this enrichment value of iMet should have higher allele frequency among virus populations. Given the mutation profile and frequency spectrum in virus populations, the only uncertainty in testing this hypothesis is that ATG does not have synonymous codon so that any changes involving ATG would either change the amino acid or even change the start codon. It is difficult to parse whether the mutation patterns are connected with the selection on the enrichment of iMet.
Regarding the enrichment of iMet, one might be confused that how could the iMet–tRNAs have prior knowledge of the number of internal ATGs in the genes when they initiate the translation by recognizing iATG? There is a potential explanation. The biological processes are essentially chemical reactions. The tRNA molecules and the mRNA molecules are mixed in the cells. When initiating the translation by recognizing iATG, although the iMet–tRNAs do not have prior knowledge of the number of internal ATGs in the genes, however, higher concentrations of iMet–tRNA over internal Met–tRNA could have an advantage of fast recognition of iATG. A greater number of internal ATGs would attract the Met–tRNAs to decode them, and as a result, Met–tRNAs have less power to compete with iMet–tRNAs to bind the iATGs. Then, translation initiation efficiency might be elevated.
There are also limitations of our work. If the higher enrichment of iMet is beneficial, then any mutations that increase this value would be favorable. These mutations include the gain of internal ATGs in the CDSs. Ideally, selection force could be inferred from the mutation spectrum. But ATG does not have synonymous codons. Any mutations that create ATG or abolish ATG would be missense mutations. This fact makes it difficult to test the mechanism proposed in this study because the mutations relevant to ATGs are subjected to selection pressure from amino acid changes. While this is a limitation, we could interpret this issue from an optimistic angle. The mutation and evolution patterns of SARS-CoV-2 are of great medical significance as knowing these messages help people prevent infection and isolate the patients. In the evolutionary studies, if researchers failed to find a reason for the effect of an ATG-related mutation, then they could consider whether this mutation affects the enrichment of iMet by altering the number of ATG codons.
Our work suggests that the genomes of SARS-CoV-2 and other viruses have the advantage of competing for the iMet–tRNAs with host mRNAs. The capture of iMet–tRNAs by start codons allows the efficient translation initiation and fast reproduction of viral RNA and proteins. The higher translation initiation efficiency might compensate the lower tAI and tRNA availability of viral genes. Our study raised a possibility of how the virus could successfully survive in the host environment, translate its own RNA, and reproduce itself from the sea of host mRNAs. In summary, if the enrichment pattern of iMet is omnipresent in a wide variety of species, then our idea could deepen people’s understanding of the host–parasite relationship, and even may help design the methods to fight against the human viruses.
Data availability
SARS-CoV-2 genome and other virus’ genomes: NCBI website (https://www.ncbi.nlm.nih.gov/genome/). The coding sequence of human genome: Ensembl website version hg19 (ftp://ftp.ensembl.org/pub/release-75/fasta/homo_sapiens/cds/). The tRNA copy number in human genome: Genomic tRNA Database (https://gtrnadb.ucsc.edu/).
Abbreviations
- SARS-CoV-2:
-
Severe Acute Respiratory Syndrome Coronavirus 2
- tRNA:
-
Transfer RNA
- tAI:
-
TRNA adaptation index
- mRNA:
-
Messenger RNA
- CDS:
-
Coding sequence
- iATG:
-
Initiation ATG
- iMet:
-
Initiation Methionine
- FDR:
-
False discovery rate
References
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300
Chu D, Wei L (2020) Reduced C-to-U RNA editing rates might play a regulatory role in stress response of Arabidopsis. J Plant Physiol 244:153081
Cowling BJ, Leung GM (2020) Epidemiological research priorities for public health control of the ongoing global novel coronavirus (2019-nCoV) outbreak. Eurosurveillance. https://doi.org/10.2807/1560-7917.ES.2020.25.6.2000110
dos Reis M, Savva R, Wernisch L (2004) Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res 32:5036–5044
Hui DS, Esam IA, Madani TA, Ntoumi F, Kock R, Dar O, Ippolito G, McHugh TD, Memish ZA, Drosten C et al (2020) The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health. The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis 91:264–266
Ikemura T, Ozeki H (1983) Codon usage and transfer RNA contents: organism-specific codon-choice patterns in reference to the isoacceptor contents. Cold Spring Harb Symp Quant Biol 47(Pt 2):1087–1097
Sabi R, Tuller T (2014) Modelling the efficiency of codon-tRNA interactions based on codon usage bias. DNA Res 21:511–526
Thompson RC, Stone PJ (1977) Proofreading of the codon-anticodon interaction on ribosomes. Proc Natl Acad Sci USA 74:198–202
Thompson RC, Dix DB, Gerson RB, Karim AM (1981) A GTPase reaction accompanying the rejection of Leu-tRNA2 by UUU-programmed ribosomes. Proofreading of the codon-anticodon interaction by ribosomes. J Biol Chem 256:81–86
Wang C, Horby PW, Hayden FG, Gao GF (2020) A novel coronavirus outbreak of global health concern. Lancet 395:470–473
Acknowledgements
We thank all the people who contributed to the world in this SARS-CoV-2 time. We also thank our group members for their support to this work.
Funding
No funding has supported this research.
Author information
Authors and Affiliations
Contributions
All authors participated in writing original draft. XW reviewed and revised this manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Stefan Hohmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, Y., Gai, Y., Li, Y. et al. SARS-CoV-2 has the advantage of competing the iMet–tRNAs with human hosts to allow efficient translation. Mol Genet Genomics 296, 113–118 (2021). https://doi.org/10.1007/s00438-020-01731-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-020-01731-4