Abstract
Strongyloides stercoralis is an intestinal helminth that infects people worldwide. Hyperinfection or disseminated human strongyloidiasis can involve vital organs, leading to lethal outcomes. We analyzed immunoproteomics of antigenic spots, derived from S. stercoralis third-stage larvae and reacted with human strongyloidiasis sera, by two-dimensional gel electrophoresis and immunoblotting. Of 26 excised antigenic spots analyzed by liquid chromatography–electrospray ionization–tandem mass spectrometry, 20 proteins were identified. Most proteins were associated with enzymes involved in the metabolic process, energy generation, and oxidation–reduction. The proteins relate to promotion of worm development, cell division, cell signaling and transportation, and regulation of muscular contraction. Identification of antigenic proteins shows promise in helping to discover potential diagnostic protein markers or vaccine candidates for S. stercoralis infection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Strongyloides stercoralis, a soil-transmitted helminth, is an intestinal roundworm that infects people worldwide (Schar et al. 2013). The life cycle of S. stercoralis is complex, including direct, auto-infective, and free-living cycles (Schar et al. 2013). Infection, resulting in human strongyloidiasis, can occur through skin penetration of infective third-stage larvae (L3) (Schar et al. 2013). It occurs primarily in poor communities, travelers and former war veterans, immigrants, immunocompromised populations, and people exposed to soil (Beknazarova et al. 2016). Some patients present gastrointestinal symptoms and hyperinfection or disseminated strongyloidiasis, which can affect several organs, leading to fatal outcomes (Grove 1996). Presently, the immunology/immunopathology, protective immune response in humans, and biology of S. stercoralis are not fully understood. This study reported the biochemical properties of antigenic protein spots derived from S. stercoralis L3 using immunoproteomic and mass spectrometry techniques. Until now, there are few reports characterizing the S. stercoralis L3 antigenic proteins by proteomic analysis (Rodpai et al. 2016; Marcilla et al. 2010). The proteomic technique is a strategy for studying the patterns of protein expression in organisms and interpreting mass spectrometry data using cross-species databases. The results could augment our knowledge of the immune response in humans, which may lead to the discovery of diagnostic protein markers or valuable vaccine candidates for helminth infection.
Materials and methods
Strongyloides stercoralis worms and protein extraction
Direct developed S. stercoralis third-stage larvae (L3) were collected from the fecal samples of infected asymptomatic patients using filter paper culture technique (Harada and Mori 1955). The L3 extract was prepared as has previously been described (Rodpai et al. 2016). The Quick Start Bradford Protein Assay (Bio-Rad Laboratories, Inc., CA) was used to detect the protein concentration.
Two-dimensional gel electrophoresis (2DE)
The L3 extract was treated with the 2-D Clean-Up Kit (GE Healthcare Bio-Sciences Corp., Piscataway, NJ). One hundred fifty micrograms of the L3 extract was resuspended in rehydration solution and loaded onto a 7-cm Immobiline DryStrip gel (IPG) with a non-linear pH gradient of 3–11 (GE Healthcare). The sample was focused according to the manufacturer’s instructions using the Ettan IPGphor 3 (GE Healthcare). Each of the focused IPG strips was then done in the second dimension on 12% SDS–polyacrylamide gel electrophoresis (SDS–PAGE). The gel was then transferred to nitrocellulose membranes for immunodetection. Another gel was also stained with colloidal Coomassie blue, and the target spots were excised for protein identification by mass spectrometry. The experiments were conducted in triplicate.
Immunodetection
The experiment was independently conducted and visualized in triplicate as has previously been described (Rodpai et al. 2016). Each of the skim milk-blocked membranes was reacted with a 1:100 diluted pool of seven human serum samples from parasitologically proven strongyloidiasis patients or a 1:100 diluted pool of seven serum samples from healthy adult volunteers without intestinal parasitic infection in their stool from non-endemic areas and then probed with a 1:10,000 diluted goat anti-human IgG (H+L) antibody, horseradish peroxidase conjugate (Invitrogen Corporation, Camarillo, CA).
Liquid chromatography–electrospray ionization–tandem mass spectrometry (LC–ESI–MS/MS) and data analysis
Coomassie-stained protein spots that matched with the reacted spots against pooled human strongyloidiasis sera but not by healthy human sera were excised from the gel using an Ettan Spot Picker (GE Healthcare Bio-Sciences AB). Each protein in-gel spots was digested with trypsin (Promega, Madison, WI). The tryptic digested samples were analyzed with LC–ESI–MS/MS. NCBI’s protein database (Metazoa) searching via the MASCOT MS/MS Ion Search program (www.matrixscience.com) was used to identify proteins. The instrument was provided by Khon Kaen University Research Instrument Center, Thailand.
The identified peptide sequences from LC–ESI–MS/MS and the MASCOT search (which generates sequences for the individual peptides) were assigned to the UniProtKB database (peptide search) (http://www.uniprot.org/) and described in gene ontology (GO) terms based on the biological process, molecular function, and cellular components. Since there was limited information regarding S. stercoralis gene sequences in the Mascot Server, peptide sequences from LC–ESI–MS/MS were subjected to the UniProtKB and WormBase ParaSite databases, in which the genome sequence of S. stercoralis is available (Hunt et al. 2016).
Results
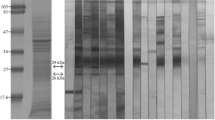
Twenty-six antigenic protein spots related to protein spots in the Coomassie-stained gel, which were recognized by human strongyloidiasis sera, are shown in Fig. 1. The target protein spots were excised from the gels and digested. The resulting peptides were then identified using mass spectrometry (Table S1) based on matches to homologous proteins from related round worms (Strongyloides ratti, Caenorhabditis remanei, Caenorhabditis elegans, and Haemonchus contortus). Twenty different proteins were identified based on matches to S. stercoralis gene sequences in the UniProtKB and WormBase ParaSite databases (Table 1 and Table S2).
Showing representative of Coomassie-stained 2DE gel and immunoblotting reaction patterns. The Strongyloides stercoralis L3 protein extract (150 μg) was separated using two-dimensional gel electrophoresis. Each colloidal Coomassie blue-stained gel (a) and the other were transferred onto a nitrocellulose membrane and then probed with human strongyloidiasis sera (b) and healthy sera (c) using immunoblotting. Coomassie-stained protein spots that matched with the reacted spots against pooled human strongyloidiasis sera were excised and subjected to LC–ESI–MS/MS. Molecular weight markers are indicated at the left of each image. Numbers and circles or arrows on images indicated the spot identity used in Table 1, Table S1, and Table S2, in which details of their identification are given
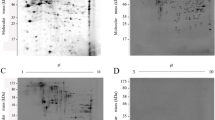
Out of a total of 20 proteins, 17 were annotated with gene ontology (GO) terms based on InterPro and QuickGO term annotations (Fig. 2 and Table S2), while three were not annotated. They were grouped according to the biological process (Fig. 2a), molecular function (Fig. 2b), and cellular component (Fig. 2c). The proteins were related to nine biological processes, 26 molecular functions, and five cellular components.
Gene ontology (GO) terms of the 20 proteins of Strongyloides stercoralis third-stage larval extract. The identified proteins were classified into biological process (a), molecular function (b), and cellular component (c) using InterPro and QuickGO. The excess numbers of total annotated proteins are because some proteins were grouped in more than one functional category
Discussion
There has already been a proteomic study of S. stercoralis antigenic protein recognized with human strongyloidiasis sera (Rodpai et al. 2016). Characterizations of the diagnostic 26- and 29-kDa polypeptide bands of S. stercoralis L3 have been reported (Rodpai et al. 2016). Here, we identified additional antigenic proteins of S. stercoralis recognized by human strongyloidiasis in patient sera using an immunoproteomic analysis.
Base on GO database comparisons, each immune-reactive protein was identified either as a cellular component, as having a molecular function, or as participating in at least one biological process. Metabolic process and energy generation, oxidation–reduction, promotion of worm development activation, cell division, cell signaling and transportation, and regulation of muscular contraction were revealed (Table S2; Fig. 2). Most proteins were associated with enzymes involved in the metabolic process and energy generation, and oxidation–reduction (16 protein spots) (Table 1), which could be used to keep the parasite alive. Five spots (nos. 18, 19, 20, 213, and 214) were identified as aconitate hydratase. It is an enzyme involved in the tricarboxylic acid cycle (Krebs cycle or the citric acid cycle) which catalyzes the reversible isomerization of citrate and isocitrate. Four spots (nos. 18, 20, 210, and 213) were identified as propionyl-CoA carboxylase alpha chain. Propionyl-CoA carboxylase is a biotin-dependent enzyme (GO:0004075), a Krebs cycle intermediate. Spot nos. 12 and 32 were identified as NADH-ubiquinone oxidoreductase or NADH dehydrogenase (mitochondrial complex I), which is a respiratory chain enzyme involved in ATP synthesis-coupled electron transport (GO:0042773) and the oxidation–reduction process (GO:0055114). These proteins are also involved in the aging of C. elegans, as RNAi knockdown of specific NADH-ubiquinone oxidoreductase has been shown to extend lifespan (Lee et al. 2003). Two spots (nos. 187 and 188) were identified as enolase involved in phosphopyruvate hydratase activity in the glycolytic process. Enolase might be used as a vaccine candidate and in immunodiagnosis, as recombinant Ascaris suum enolase can immunize mice to responses against A. suum infection (Chen et al. 2012; Wang et al. 2012). One spot (no. 180) was identified as a medium-chain specific acyl-CoA dehydrogenase enzyme possibly involved in the metabolism of fatty acids, as has been reported in A. suum (Komuniecki et al. 1985). One spot (no. 36) was identified as belonging to the acetyl-CoA hydrolase/transferase family. The acetyl-CoA hydrolase/transferase C-terminal domain-containing enzyme participates in pyruvate metabolism (GO:0006084). Two spots (nos. 179 and 180) were identified as citrate synthase, a key enzyme to catalyze the first reaction of the citric acid cycle (GO:0006099) that catalyzes the condensation of oxaloacetate and acetyl-CoA to form citrate. One spot (no. 154) was identified as arginine kinase F46H5.3, which belongs to the ATP:guanido phosphotransferase family. The structure and function of this group are related to enzymes that involve the transfer of phosphate between ATP and various phosphogens. One spot (no. 146) was identified as fructose-bisphosphate aldolase 1. This enzyme functions in gluconeogenesis and glycolysis pathways (GO:0006096), which are related to a reversible reaction of the aldol and fructose 1,6-bisphosphate. Four of these metabolic proteins (propionyl-CoA carboxylase alpha chain, enolase, citrate synthase, and arginine kinase) have been identified in S. stercoralis L3 using proteomic analysis approaches (Marcilla et al. 2010). Moreover, these proteins have also been identified as being immunoreactive in nematode infection, such as that of Trichostrongylus colubriformis (Kiel et al. 2007).
The metabolic reactions, i.e., enzymes related to catabolism of lipid storage in S. stercoralis L3, are relatively understudied, despite their potential use in the design of enzymatic inhibitors that could provide novel chemotherapeutic interventions or drug targets. Metabolic pathways are essential for an organism’s survival, which require extract energy from carbon compounds. Therefore, inhibition of enzymes in these pathways likely shows a good choice of therapeutic strategy to resist parasitic infections (Timson 2016).
Two spots (nos. 136 and 140) were identified as galectin, a protein which binds sugars with a specific affinity for β-galactoside. It has a variety of mediation functions including transmembrane signaling, cell–cell interactions, and cell–matrix adhesion. This protein is related to initiating host immune response and has implications for vaccine development (Vasta 2009).
One spot (no. 138) was identified as a guanine nucleotide-binding protein (G protein) subunit beta-4, which mediates the chemosensory signals that regulate S. stercoralis L3i activation in cyclic guanosine monophosphate (cGMP) pathway signaling. This G-beta subunit gene was expressed in all developmental stages (Stoltzfus et al. 2014).
Twelve immune reactive spots were identified as the locomotion protein components of the cytoskeleton in the nematode (Ono 2014) (i.e., actin, paramyosin and myosin heavy chain, troponin complex, and calponin repeat-containing protein). Three spots (nos. 10, 11, and 177) were identified as actin, the protein associated with muscle contractility in C. elegans (Ono 2014). Two spots (nos. 25 and 206) were identified as paramyosin and myosin heavy chain proteins, which are known for their role in muscle contraction in the nematode (Kagawa et al. 2007). Six spots (nos. 25, 36, 179, 180, 187, and 188) were identified as calponin repeat-containing protein. Calponin is a cytoskeletal protein that regulates actomyosin contractility and stabilizes actin filaments (Rozenblum and Gimona 2008). Identified immunoreactive spots (nos. 139, 140, and 206) are related to troponin T and troponin I, the tropomyosin subunits. The troponin complex is a heteromeric protein attached to tropomyosin that regulates muscle contraction in the nematode C. elegans (Kagawa et al. 2007). Tropomyosin, an invertebrate pan-allergen, has been reported to be a helminthic allergen, such as Anisakis simplex, that occurs in response to high IgE titer in asymptomatic Anisakis-infected patients (Asturias et al. 2000). The structural muscle proteins such as myosin, paramyosin, and calponin of T. colubriformis have been identified as immunoreactive in immune sheep (Kiel et al. 2007). Interestingly, the cytoskeletal proteins (actin, paramyosin, myosin heavy chain, and tropomyosin) have been identified as the most abundant transcripts encoding predicted excretory–secretory proteins of S. stercoralis L3, which are likely related to host–parasite interaction and host immune system regulation (Marcilla et al. 2012). One spot (no. 2) was identified as 14-3-3 protein zeta. The 14-3-3 proteins are acidic proteins, a large family of broadly expressed 24- to 33-kDa proteins, with a role in regulatory signal transmission related to cell migration, proliferation, and morphology changes which occur during the parasite life cycle (Siles-Lucas Mdel and Gottstein 2003). These proteins also are potential good targets for anthelmintic drugs and vaccines (Siles-Lucas Mdel and Gottstein 2003) and for diagnostic method development (Rodpai et al. 2016).
Among the 26 identified spots, we found that 10 spots (38.46%) contained multiple proteins (Table 1 and Table S2). Multiple proteins were likely detected in a single spot because of the high sensitivity of protein identification by LC–ESI–MS/MS (Lim et al. 2003) and the limited separation performance of 2D gels. We also found that 10 proteins (50%) of the 20 identified proteins were identified in multiple spots (Table 1 and Table S2). In this case, multiple spots indicated either conformational changes that result in segregation of proteins to different isoelectric points (Deng et al. 2012) or the presence of protein isoforms (Kiel et al. 2007).
This study suggests that a relatively large number of parasite antigens are involved in immune responses to S. stercoralis infection and helpful in understanding the molecular mechanisms that support host–parasite interaction. The identified antigenic proteins could possibly help in developing a strategy for the mass production of antigens using a recombinant technique. These could, in turn, be used in the development of immunodiagnostic tests for human strongyloidiasis. Nevertheless, the limitation of this experiment is that the entire smeary signal on the western blots is specific for Strongyloides patient sera possibly due to the nature of the Strongyloides L3 extract, i.e., glycoproteins, in spite of the purification through the 2-D Clean-Up Kit. However, next experiments need to be used for clarifications.
References
Asturias JA, Eraso E, Moneo I, Martínez A (2000) Is tropomyosin an allergen in Anisakis? Allergy 55:898–899
Beknazarova M, Whiley H, Ross K (2016) Strongyloidiasis: a disease of socioeconomic disadvantage. Int J Environ Res Public Health 13:E517. doi:10.3390/ijerph13050517
Chen N, Yuan ZG, Xu MJ, Zhou DH, Zhang XX, Zhang YZ, Wang XW, Yan C, Lin RQ, Zhu XQ (2012) Ascaris suum enolase is a potential vaccine candidate against ascariasis. Vaccine 30:3478–3482. doi:10.1016/j.vaccine.2012.02.075
Deng X, Hahne T, Schroder S, Redweik S, Nebija D, Schmidt H, Janssen O, Lachmann B, Watzig H (2012) The challenge to quantify proteins with charge trains due to isoforms or conformers. Electrophoresis 33:263–269
Grove DI (1996) Human strongyloidiasis. Adv Parasitol 38:251–309
Harada U, Mori OA (1955) A new method for culturing hookworm. Yonago Acta Med 1:177–179
Hunt VL, Tsai IJ, Coghlan A et al (2016) The genomic basis of parasitism in the Strongyloides clade of nematodes. Nat Genet 48:299–307. doi:10.1038/ng.3495
Kagawa H, Takaya T, Ruksana R, Anokye-Danso F, Amin MZ, Terami H (2007) C. elegans model for studying tropomyosin and troponin regulations of muscle contraction and animal behavior. Adv Exp Med Biol 592:153–161. doi:10.1007/978-4-431-38453-3_14
Kiel M, Josh P, Jones A, Windon R, Hunt P, Kongsuwan K (2007) Identification of immuno-reactive proteins from a sheep gastrointestinal nematode, Trichostrongylus colubriformis, using two-dimensional electrophoresis and mass spectrometry. Int J Parasitol 37:1419–1429. doi:10.1016/j.ijpara.2007.04.016
Komuniecki R, Fekete S, Thissen-Parra J (1985) Purification and characterization of the 2-methyl branched-chain acyl-CoA dehydrogenase, an enzyme involved in NADH-dependent enoyl-CoA reduction in anaerobic mitochondria of the nematode, Ascaris suum. J Biol Chem 260:4770–4777
Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G (2003) A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet 33:40–48. doi:10.1038/ng1056
Lim H, Eng J, Yates JR 3rd, Tollaksen SL, Giometti CS, Holden JF, Adams MW, Reich CI, Olsen GJ, Hays LG (2003) Identification of 2D-gel proteins: a comparison of MALDI/TOF peptide mass mapping to mu LC-ESI tandem mass spectrometry. J Am Soc Mass Spectrom 14:957–970
Marcilla A, Sotillo J, Perez-Garcia A, Igual-Adell R, Valero ML, Sanchez-Pino MM, Bernal D, Munoz-Antoli C, Trelis M, Toledo R, Esteban JG (2010) Proteomic analysis of Strongyloides stercoralis L3 larvae. Parasitology 137:1577–1583
Marcilla A, Garg G, Bernal D, Ranganathan S, Forment J, Ortiz J, Munoz-Antoli C, Dominguez MV, Pedrola L, Martinez-Blanch J, Sotillo J, Trelis M, Toledo R, Esteban JG (2012) The transcriptome analysis of Strongyloides stercoralis L3i larvae reveals targets for intervention in a neglected disease. PLoS Negl Trop Dis 6:e1513. doi:10.1371/journal.pntd.0001513
Ono S (2014) Regulation of structure and function of sarcomeric actin filaments in striated muscle of the nematode Caenorhabditis elegans. Anat Rec 297:1548–1559. doi:10.1002/ar.22965
Rodpai R, Intapan PM, Thanchomnang T, Sanpool O, Janwan P, Laummaunwai P, Wongkham C, Insawang T, Maleewong W (2016) Strongyloides stercoralis diagnostic polypeptides for human strongyloidiasis and their proteomic analysis. Parasitol Res 115:4007–4012. doi:10.1007/s00436-016-5170-7
Rozenblum GT, Gimona M (2008) Calponins: adaptable modular regulators of the actin cytoskeleton. Int J Biochem Cell Biol 40:1990–1995. doi:10.1016/j.biocel.2007.07.010
Schar F, Trostdorf U, Giardina F, Khieu V, Muth S, Marti H, Vounatsou P, Odermatt P (2013) Strongyloides stercoralis: global distribution and risk factors. PLoS Negl Trop Dis 7:e2288. doi:10.1371/journal.pntd.0002288
Siles-Lucas Mdel M, Gottstein B (2003) The 14-3-3 protein: a key molecule in parasites as in other organisms. Trends Parasitol 19:575–581
Stoltzfus JD, Bart SM, Lok JB (2014) cGMP and NHR signaling co-regulate expression of insulin-like peptides and developmental activation of infective larvae in Strongyloides stercoralis. PLoS Pathog 10:e1004235. doi:10.1371/journal.ppat.1004235
Timson DJ (2016) Metabolic enzymes of helminth parasites: potential as drug targets. Curr Protein Pept Sci 17:280–295
Vasta GR (2009) Roles of galectins in infection. Nat Rev Microbiol 7:424–438
Wang Y, Zhang J, Yuan ZY, Zhou HJ, Shen YJ, Xu YX, Wang YJ, Wu WP, Cao JP (2012) Cloning, expression and immunodiagnostic evaluation of enolase from Echinococcus granulosus. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi 24:549–552 556
Acknowledgements
We wish to acknowledge the support of the English Consultation Clinic at the Khon Kaen University Faculty of Medicine Research Affairs Division and the Khon Kaen University Publication Clinic at the Research and Technology Transfer Affairs Division for their assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was supported by a TRF Senior Research Scholar Grant, Thailand Research Fund grant number RTA5880001; the Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission, Thailand, through the Health Cluster (SHeP-GMS); and the Faculty of Medicine, Khon Kaen University (grant number TR57201) through WM and PMI. RR was supported by the Thailand Research Fund through the Royal Golden Jubilee Ph.D. Program (grant no. PHD/0053/2556). OS was supported by a scholarship under the Post-Doctoral Training Program from the Research Affairs and Graduate School, Khon Kaen University (grant no. 58101).
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Khon Kaen University Ethics Committee for Human Research (HE591192) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Rights and permissions
About this article
Cite this article
Rodpai, R., Intapan, P.M., Thanchomnang, T. et al. Identification of antigenic proteins in Strongyloides stercoralis by proteomic analysis. Parasitol Res 116, 1687–1693 (2017). https://doi.org/10.1007/s00436-017-5443-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-017-5443-9