Abstract
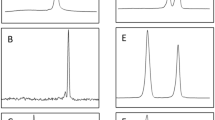
The microchip-based capillary electrophoresis technology represents a valuable recent development for the analysis of complex DNA banding patterns. We have used this technology for the differentiation of the closely related cyathostomin species Cylicocyclus elongatus and C. insigne from the horse. We found that the Agilent 2100 bioanalyser in combination with the DNA 7500 Lab Chip were suited to perform a phylogenetic DNA fingerprinting analysis of the parasite species studied. The analysis of the electrophoretic data was optimised and it was possible to resolve a phylogenetic tree where all 12 individual worms of the two Cylicocyclus species studied were assigned to their species as determined by microscopic identification based on morphological traits. Thus, our data indicated that the procedure described here provides an additional powerful tool that can be employed for species delineation of closely related strains or species, such as the two taxa of Cylicocyclus investigated in the present study. Furthermore, by determining the second internal transcribed spacer region of three and nine individual worms for C. elongatus and C. insigne, respectively, low intraspecific variations of only up to 0.3% were demonstrated.






Similar content being viewed by others

References
Adamson RE, Ward RD, Feliciangeli MD, Maingon R (1993) The application of random amplified polymorphic DNA for sandfly species identification. Med Vet Entomol 7:203–207
Bucknell DG, Gasser RB, Beveridge I (1995) The prevalence and epidemiology of gastrointestinal parasites of horses in Victoria, Australia. Int J Parasitol 25:711–724
Burr MD, Pepper IL (1997) Variability in presence-absence scoring of AP PCR fingerprints affects computer matching of bacterial isolates. J Microbiol Methods 29:63–68
Chilton NB, Gasser RB, Beveridge I (1997) Phylogenetic relationships of Australian strongyloid nematodes inferred from ribosomal DNA sequence data. Int J Parasitol 27:1481–1494
Costa CA, Gomes RF, Melo MN, Ribeiro MF (2001) Eimeria parasites of domestic fowl: genetic relationships of different isolates estimated from random amplified polymorphic DNA. Parasitol Res 87:459–466
Dias NE, de Souza CP, Rollinson D, Katz N, Pena SD, Simpson AJ (1993) The random amplification of polymorphic DNA allows the identification of strains and species of schistosome. Mol Biochem Parasitol 57:83–88
Dolnik V, Liu S, Jovanovich S (2000) Capillary electrophoresis on microchip. Electrophoresis 21:41–54
Epe C, Bienioschek S, Rehbein S, Schnieder T (1995) Comparative RAPD-PCR analysis of lungworms (Dictyocaulidae) from fallow deer, cattle, sheep, and horses. Zentralbl Veterinaermed Reihe B 42:187–191
Epe C, Meuwissen M, Stoye M, Schnieder T (1999) Transmission trials, ITS-2-PCR and RAPD-PCR show identity of Toxocara canis isolates from red fox and dog. Vet Parasitol 84:101–112
Gasser RB, Monti JR (1997) Identification of parasitic nematodes by PCR-SSCP of ITS-2 rDNA. Mol Cell Probes 11:201–209
Gasser RB, Stevenson LA, Chilton NB, Nansen P, Bucknell DG, Beveridge I (1996) Species markers for equine strongyles detected in intergenic rDNA by PCR-RFLP. Mol Cell Probes 10:371–378
Georgi JR (1980) Parasitology for veterinarians, 3rd edn. Saunders, Philadelphia, Pa.
Hampl V, Pavlicek A, Flegr J (2001) Construction and bootstrap analysis of DNA fingerprinting-based phylogenetic trees with the freeware program FreeTree: application to trichomonad parasites. Int J Syst Evol Microbiol 51:731–735
Humbert JF, Cabaret J (1995) Use of random amplified polymorphic DNA for identification of ruminant trichostrongylid nematodes. Parasitol Res 81:1–5
Hung GC, Chilton NB, Beveridge I, McDonnell A, Lichtenfels JR, Gasser RB (1997) Molecular delineation of Cylicocyclus nassatus and C. ashworthi (Nematoda:Strongylidae). Int J Parasitol 27:601–605
Hung GC, Chilton NB, Beveridge I, Gasser RB (1999) Secondary structure model for the ITS-2 precursor rRNA of strongyloid nematodes of equids: implications for phylogenetic inference. Int J Parasitol 29:1949–1964
Hung GC, Chilton NB, Beveridge I, Gasser RB (2000) A molecular systematic framework for equine strongyles based on ribosomal DNA sequence data. Int J Parasitol 30:95–103
Leignel V, Humbert JF, Elard L (1997) Study by ribosomal DNA ITS 2 sequencing and RAPD analysis on the systematics of four Metastrongylus species (Nematoda:Metastrongyloidea). J Parasitol 83:606–611
Lichtenfels JR (1975) Helminths of domestic equids. Illustrated keys to genera and species with emphasis on North American forms. Proc Helminth Soc Wash 42:1–92
Lichtenfels JR, Kharchenko VA, Sommer C, Ito M (1997) key characters for the microscopical identification of Cylicocyclus nassatus and Cylicocyclus ashworthi (Nematoda: Cyathostominae) of the horse, Equus caballus. J Helminthol Soc Wash 64:120–127
Lichtenfels JR, Gibbons LM, Krecek RC (2002) Recommended terminology and advances in the systematics of Cyathostominae (Nematoda: Strongyloidea) of horses. Vet Parasitol 107:337–342
Lu C-Y, Tso D-J, Yang T, Jong Y-J, Wie Y-H (2002) Detection of DNA mutations associated with mitochondrial diseases by Agilent 2100 bioanalyzer. Clin Chim Acta 318:97–105
Lyons ET, Tolliver SC, Drudge JH (1999) Historical perspective of cyathostomes: prevalence, treatment and control programs. Vet Parasitol 85:97–111
McDonnell A, Love S, Tait A, Lichtenfels JR, Matthews JB (2000) Phylogenetic analysis of partial mitochondrial cytochrome oxidase c subunit I and large ribosomal RNA sequences and nuclear internal transcribed spacer I sequences from species of Cyathostominae and Strongylinae (Nematoda, order Strongylida), parasites of the horse. Parasitology 121:649–659
Nachamkin I, Panaro NJ, Li M, Ung H, Yuen PK, Kricka LJ, Wilding P (2001) Agilent 2100 bioanalyzer for restriction fragment length polymorphism analysis of the Campylobacter jejuni flagellin gene. J Clin Microbiol 39:754–757
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Nylander JAA (2002) Testing models of evolution—MrModeltest version 1.1b. Department of Systematic Zoology, Evolutionary Biology Centre, Uppsala University, Uppsala
Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Panaro NJ, Yuen PK, Sakazume T, Fortina P, Kricka LJ, Wilding P (2000) Evaluation of DNA fragment sizing and quantification by the agilent 2100 bioanalyzer. Clin Chem 46:1851–1853
Pavlicek A, Hrda S, Flegr J (1999) Free-Tree—freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness. Application in the RAPD analysis of genus Frenkelia. Folia Biol (Prague) 45:97–99
Procunier JD, Fernando MA, Barta JR (1993) Species and strain differentiation of Eimeria spp. of the domestic fowl using DNA polymorphisms amplified by arbitrary primers. Parasitol Res 79:98–102
Sciacchitano CJ (1998) DNA fingerprinting of Listeria monocytogenes using enterobacterial repetitive intergenic consensus (ERIC) motifs-polymerase chain reaction/capillary electrophoresis. Electrophoresis 19:66–70
Siles-Lucas M, Felleisen R, Cuesta-Bandera C, Gottstein B, Eckert J (1994) Comparative genetic analysis of Swiss and Spanish isolates of Echinococcus granulosus by southern hybridization and random amplified polymorphic DNA technique. Appl Parasitol 35:107–117
Steindel M, Dias NE, de Menezes CL, Romanha AJ, Simpson AJ (1993) Random amplified polymorphic DNA analysis of Trypanosoma cruzi strains. Mol Biochem Parasitol 60:71–79
Swofford DL (1998) PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sinauer, Sunderland, Mass.
Troell K, Mattsson JG, Alderborn A, Höglund J (2003) Pyrosequencing analysis identifies discrete populations of Haemonchus contortus from small ruminants. Int J Parasitol 33:765–771
Valentini A, Timperio AM, Cappuccio I, Zolla L (1996) Random amplified polymorphic DNA (RAPD) interpretation requires a sensitive method for the detection of amplified DNA. Electrophoresis 17:1553–1554
Welsh J, McClelland M (1990) Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res 18:7213–7218
Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Acknowledgements
The work reported here was supported by a grant from the Deutsche Forschungsgemeinschaft (Schn. 267/13–1). The authors wish to thank Dr Judith McAlister-Hermann and Dr William Blackhall for help in revising the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Posedi, J., Drögemüller, M., Schnieder, T. et al. Microchip capillary electrophoresis-based genetic comparison of closely related cyathostomin nematode parasites of horses using randomly amplified polymorphic DNA polymerase chain reaction. Parasitol Res 92, 421–429 (2004). https://doi.org/10.1007/s00436-003-1067-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-003-1067-3