Abstract
Purpose
Necroptosis plays an essential role in oncogenesis and tumor progression in hepatocellular carcinoma (HCC). This study aimed to investigate the role of necroptosis in the development and progression of HCC. Specifically, we constructed a prognostic prediction model using necroptosis-associated genes (NAGs) to predict patient outcomes.
Methods
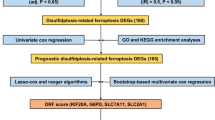
Using data from The Cancer Genome Atlas (TCGA) database, we analyzed gene expression and clinical data. We identified a 5-gene model associated with NAGs and explored genetic features and immune cell infiltration using the CIBERSORT algorithm. In addition, we conducted single-cell RNA sequencing to investigate the potential role of necroptosis in HCC.
Results
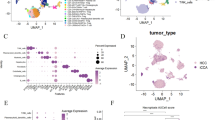
We constructed a 5-gene prognostic model based on NAGs that demonstrated excellent predictive accuracy in both training and validation sets. Using multifactorial cox regression analysis, we confirmed the risk score derived from the model as an independent predictor of prognosis, surpassing other clinical characteristics. Patients with high risk scores had significantly worse prognosis than those with low risk scores. To enhance the clinical utility of the necroptosis score, we constructed an accurate nomogram. Additionally, we compared metabolic pathway and immune microenvironment differences between HCC tumors with high and low risk scores. Our single-cell RNA sequencing analyses revealed that necroptosis in HCC was primarily associated with a specific subset of macrophages.
Conclusions
Our study revealed the presence of two distinct necroptosis subtypes in HCC and developed a robust prognostic model with exceptional predictive accuracy. We observed significantly higher infiltration of M0 macrophages in the high-risk group. We propose that rescuing cytochrome c metabolism in HCC could serve as a potential therapeutic strategy. Furthermore, at a single-cell resolution, our analysis identified myeloid cells as the primary cells exhibiting necroptosis. Specifically, macrophages expressing CD5L, CETP, and MARCO, which may belong to a subset of tissue-resident macrophages, were found to be highly susceptible to necroptosis. These findings suggest the involvement of this specific macrophage subset in potential antitumor therapies. Our study provides novel insights into predicting patient prognosis and developing personalized therapeutic approaches for HCC.





Similar content being viewed by others
Data availability
The TCGA-LIHC dataset generated during the current study are available in the UCSC Xena (https://xena.ucsc.edu). The ScRNA-seq datasets generated during the current study are available in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE149614).
Abbreviations
- single-cell RNA seq:
-
Single-cell RNA sequencing
- bulk RNA seq:
-
Bulk RNA sequencing
- HCC:
-
Hepatocellular carcinoma
- iCCA/ICC:
-
Intrahepatic cholangiocarcinoma
- NAGs:
-
Necroptosis associated genes
- TCGA:
-
The Cancer Genome Atlas
- LASSO:
-
Least absolute shrinkage and selection operator
- AIC:
-
Akaike information criterion
- GSEA:
-
Gene Set Enrichment Analysis
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- KM:
-
Kaplan–Meier
- PCA:
-
Principal Components Analysis
- ROC:
-
Receiver Operating Characteristic curve
- AUC:
-
Area under curve
- NES:
-
Normalized Enrichment Score
- OS:
-
Overall survival
- DEGs:
-
Differential expression genes
- TRMs:
-
Tissue-resident macrophages
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL et al (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424. https://doi.org/10.3322/caac.21492
Cai Z, Zhang A, Choksi S, Li W et al (2016) Activation of cell-surface proteases promotes necroptosis, inflammation and cell migration. Cell Res 26(8):886–900. https://doi.org/10.1038/cr.2016.87
Cai D, Zhao Z, Hu J, Dai X et al (2022) Identification of the tumor immune microenvironment and therapeutic biomarkers by a novel molecular subtype based on aging-related genes in hepatocellular carcinoma. Front Surg 9:836080. https://doi.org/10.3389/fsurg.2022.836080
Cheng J, Qian D, Ding X, Song T et al (2018) High PGAM5 expression induces chemoresistance by enhancing Bcl-xL-mediated anti-apoptotic signaling and predicts poor prognosis in hepatocellular carcinoma patients. Cell Death Dis 9(10):991. https://doi.org/10.1038/s41419-018-1017-8
Dick SA, Wong A, Hamidzada H, Nejat S et al (2022) Three tissue resident macrophage subsets coexist across organs with conserved origins and life cycles. Sci Immunol 7(67):eabf7777. https://doi.org/10.1126/sciimmunol.abf7777
Ding F, Li J, Zhang Y, Wang C et al (2022) Identifying a novel endoplasmic reticulum-related prognostic model for hepatocellular carcinomas. Oxid Med Cell Longev 2022:8248355. https://doi.org/10.1155/2022/8248355
Dong Q, Zhang S, Zhang H, Sun J et al (2023) MARCO is a potential prognostic and immunotherapy biomarker. Int Immunopharmacol 116:109783. https://doi.org/10.1016/j.intimp.2023.109783
Dorighello GG, Assis LHP, Rentz T, Morari J et al (2022) Novel role of CETP in macrophages: reduction of mitochondrial oxidants production and modulation of cell immune-metabolic profile. Antioxidants (Basel). https://doi.org/10.3390/antiox11091734
Gravitz L (2014) Liver cancer. Nature 516(7529):S1. https://doi.org/10.1038/516S1a
Huang R, Liu J, Li H, Zheng L et al (2021) Identification of hub genes and their correlation with immune infiltration cells in hepatocellular carcinoma based on GEO and TCGA databases. Front Genet 12:647353. https://doi.org/10.3389/fgene.2021.647353
Jiao D, Cai Z, Choksi S, Ma D et al (2018) Necroptosis of tumor cells leads to tumor necrosis and promotes tumor metastasis. Cell Res 28(8):868–870. https://doi.org/10.1038/s41422-018-0058-y
La Fleur L, Botling J, He F, Pelicano C et al (2021) Targeting MARCO and IL37R on immunosuppressive macrophages in lung cancer blocks regulatory T cells and supports cytotoxic lymphocyte function. Cancer Res 81(4):956–967. https://doi.org/10.1158/0008-5472.CAN-20-1885
Li L, Wang H (2016) Heterogeneity of liver cancer and personalized therapy. Cancer Lett 379(2):191–197. https://doi.org/10.1016/j.canlet.2015.07.018
Li D, Li K, Zhang W, Yang KW et al (2022) The m6A/m5C/m1A regulated gene signature predicts the prognosis and correlates with the immune status of hepatocellular carcinoma. Front Immunol 13:918140. https://doi.org/10.3389/fimmu.2022.918140
Long J, Wang A, Bai Y, Lin J et al (2019) Development and validation of a TP53-associated immune prognostic model for hepatocellular carcinoma. EBioMedicine 42:363–374. https://doi.org/10.1016/j.ebiom.2019.03.022
Lu Y, Yang A, Quan C, Pan Y et al (2022) A single-cell atlas of the multicellular ecosystem of primary and metastatic hepatocellular carcinoma. Nat Commun 13(1):4594. https://doi.org/10.1038/s41467-022-32283-3
Marquardt JU, Andersen JB, Thorgeirsson SS (2015) Functional and genetic deconstruction of the cellular origin in liver cancer. Nat Rev Cancer 15(11):653–667. https://doi.org/10.1038/nrc4017
Najafov A, Chen H, Yuan J (2017) Necroptosis and cancer. Trends Cancer 3(4):294–301. https://doi.org/10.1016/j.trecan.2017.03.002
Sanjurjo L, Aran G, Tellez E, Amezaga N et al (2018) CD5L promotes M2 macrophage polarization through autophagy-mediated upregulation of ID3. Front Immunol 9:480. https://doi.org/10.3389/fimmu.2018.00480
Santana KG, Righetti RF, Breda CNS, Dominguez-Amorocho OA et al (2021) Cholesterol-ester transfer protein alters M1 and M2 macrophage polarization and worsens experimental elastase-induced pulmonary emphysema. Front Immunol 12:684076. https://doi.org/10.3389/fimmu.2021.684076
Schwabe RF, Luedde T (2018) Apoptosis and necroptosis in the liver: a matter of life and death. Nat Rev Gastroenterol Hepatol 15(12):738–752. https://doi.org/10.1038/s41575-018-0065-y
Seehawer M, Heinzmann F, D’Artista L, Harbig J et al (2018) Necroptosis microenvironment directs lineage commitment in liver cancer. Nature 562(7725):69–75. https://doi.org/10.1038/s41586-018-0519-y
Sia D, Villanueva A, Friedman SL, Llovet JM (2017) Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology 152(4):745–761. https://doi.org/10.1053/j.gastro.2016.11.048
Sprooten J, De Wijngaert P, Vanmeerbeerk I, Martin S et al (2020) Necroptosis in immuno-oncology and cancer immunotherapy. Cells. https://doi.org/10.3390/cells9081823
Strilic B, Yang L, Albarran-Juarez J, Wachsmuth L et al (2016) Tumour-cell-induced endothelial cell necroptosis via death receptor 6 promotes metastasis. Nature 536(7615):215–218. https://doi.org/10.1038/nature19076
Sun L, Wang H, Wang Z, He S et al (2012) Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148(1–2):213–227. https://doi.org/10.1016/j.cell.2011.11.031
Tang B, Zhu J, Zhao Z, Lu C et al (2021) Diagnosis and prognosis models for hepatocellular carcinoma patient’s management based on tumor mutation burden. J Adv Res 33:153–165. https://doi.org/10.1016/j.jare.2021.01.018
Wang W, Lu J, Zhu F, Wei J et al (2012) Pro-apoptotic and anti-proliferative effects of mitofusin-2 via Bax signaling in hepatocellular carcinoma cells. Med Oncol 29(1):70–76. https://doi.org/10.1007/s12032-010-9779-6
Watanabe Y, Fukuda T, Hayashi C, Nakao Y et al (2022) Extracellular vesicles derived from GMSCs stimulated with TNF-alpha and IFN-alpha promote M2 macrophage polarization via enhanced CD73 and CD5L expression. Sci Rep 12(1):13344. https://doi.org/10.1038/s41598-022-17692-0
Xu Z, Peng B, Liang Q, Chen X et al (2021) Construction of a ferroptosis-related nine-lncRNA signature for predicting prognosis and immune response in hepatocellular carcinoma. Front Immunol 12:719175. https://doi.org/10.3389/fimmu.2021.719175
Zhou C, Lu X, Wang Y, Qian X et al (2022) Histopathological components correlated with MRI features and prognosis in. Eur Radiol 32(10):6702–6711
Acknowledgements
We thank all the team members for their hard work and efforts.
Funding
This study was supported by Shenzhen Science and Technology Program (Grant No. JSGG20201102162802008) and Key clinical research projects of Peking University Shenzhen Hospital (LCYJZD2021007).
Author information
Authors and Affiliations
Contributions
CJ was the study’s designer and director and wrote the manuscript. WC designed the study, carried out the bioinformatics analysis and data generation, and wrote the initial manuscript. TL and CC contributed to writing, reviewing, editing, and obtaining funding. CY conducted the literature collection. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Informed consent
Not applicable.
Publication consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, J., Chen, C., Tao, L. et al. A comprehensive analysis of the potential role of necroptosis in hepatocellular carcinoma using single-cell RNA Seq and bulk RNA Seq. J Cancer Res Clin Oncol 149, 13841–13853 (2023). https://doi.org/10.1007/s00432-023-05208-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-023-05208-w