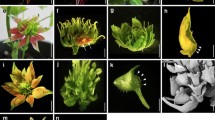
Abstract
Wintersweet (Chimonanthus praecox), a basal angiosperm endemic to China, has high ornamental value for developing beautiful flowers with strong fragrance. The molecular mechanism regulating flower development in wintersweet remains largely elusive. In this project, we seek to determine the molecular features and expression patterns of the C. praecox paleoAP3-type gene CpAP3 and examine its potential role in regulating floral development via ectopic expression in Arabidopsis thaliana and Petunia hybrida. The expression of CpAP3 is tissue-specific, with the highest level in the tepals, moderate level in carpels, and weak levels in stamen and vegetative stem tissues. Its dynamic expression during flowering is associated with flower-bud formation. Ectopic expression of CpAP3 partially rescued stamen development in ap3 mutant Arabidopsis. Although no phenotypic effect has been observed in wild-type Arabidopsis, CpAP3 overexpression in petunia brought rich morphological changes and homeotic conversions to flowers, mainly involving disruption of petal and stamen development. Expressed in a broader range than those canonical B-function regulators, the ancestral B-class gene CpAP3 can affect petal and stamen development in higher eudicots. This gene also holds some bioengineering potential in creating novel floral germplasms.






Similar content being viewed by others
Abbreviations
- MADS:
-
Yeast MCM1, Arabidopsis AGAMOUS, snapdragon DEFICIENS A and human SRF
- AP3:
-
APETALA3
- PI:
-
PISTILLATA
- Mya:
-
Million years ago
- TM6:
-
TOMATO MADS-BOX GENE6
- cDNA:
-
Complementary DNA
- RACE:
-
Rapid amplification of cDNA ends
- bp:
-
Base pairs
- ORF:
-
Open reading frame
- RT-PCR:
-
Reverse transcription-PCR
- CaMV:
-
Cauliflower mosaic virus
References
Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RJ (2000) Molecular and genetic analyses of the Silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol Cell 5:569–579
Angenent GC, Franken J, Busscher M, Weiss D, van Tunen AJ (1994) Co-suppression of the petunia homeotic gene fbp2 affects the identity of the generative meristem. Plant J 5:33–44
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353:31–37
de Martino G, Pan I, Emmanuel E, Levy A, Irish VF (2006) Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell 18:1833–1845
Eckardt NA (2006) Functional divergence of AP3 genes in the MAD world of flower development. Plant Cell 18:1779–1781
Halfter U, Ali N, Stockhaus J, Ren L, Chua NH (1994) Ectopic expression of a single homeotic gene, the Petunia gene green petal, is sufficient to convert sepals to petaloid organs. EMBO J 13:1443–1449
Hernández-Hernández T, Martínez-Castilla LP, Alvarez-Buylla ER (2007) Functional diversification of B MADS-box homeotic regulators of flower development: adaptive evolution in protein–protein interaction domains after major gene duplication events. Mol Biol Evol 24:465–481
Honma T, Goto K (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409:525–529
Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231
Irish VF (2003) The evolution of floral homeotic gene function. Bioessays 25:637–646
Jack T, Brockman LL, Meyerowitz EM (1992) The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68:683–697
Jack T, Fox GL, Meyerowitz EM (1994) Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76:703–716
Kanno A, Saeki H, Kameya T, Saedler H, Theissen G (2003) Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana). Plant Mol Biol 52:831–841
Kapoor M, Tsuda S, Tanaka Y, Mayama T, Okuyama Y, Tsuchimoto S, Takatsuji H (2002) Role of petunia pMADS3 in determination of floral organ and meristem identity, as revealed by its loss of function. Plant J 32:115–127
Kater MM, Colombo L, Franken J, Busscher M, Masiero S, Van Lookeren Campagne MM, Angenent GC (1998) Multiple AGAMOUS homologs from cucumber and petunia differ in their ability to induce reproductive organ fate. Plant Cell 10:171–182
Kim S, Koh J, Yoo MJ, Kong H, Hu Y, Ma H, Soltis PS, Soltis DE (2005) Expression of floral MADS-box genes in basal angiosperms: implications for the evolution of floral regulators. Plant J 43:724–744
Kramer EM, Dorit RL, Irish VF (1998) Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149:765–783
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5:150–163
Liu C, Zhang J, Zhang N, Shan H, Su K, Zhang J, Meng Z, Kong H, Chen Z (2010) Interactions among proteins of floral MADS-box genes in basal eudicots: implications for evolution of the regulatory network for flower development. Mol Biol Evol 27:1598–1611
Melzer R, Wang YQ, Theissen G (2010) The naked and the dead: the ABCs of gymnosperm reproduction and the origin of the angiosperm flower. Semin Cell Dev Biol 21:118–128
Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y (2003) SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130:705–718
Nicely KA (1965) A monographic study of the Calycanthaceae. Castanea 30:38–81
Piwarzyk E, Yang Y, Jack T (2007) Conserved C-terminal motifs of the Arabidopsis proteins APETALA3 and PISTILLATA are dispensable for floral organ identity function. Plant Physiol 145:1495–1505
Pnueli L, Abu-Abeid M, Zamir D, Nacken W, Schwarz-Sommer Z, Lifshitz E (1991) The MADS box gene family in tomato: temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J 1:255–266
Poupin MJ, Federici F, Medina C, Matus JT, Timmermann T, Arce-Johnson P (2007) Isolation of the three grape sub-lineages of B-class MADS-box TM6, PISTILLATA and APETALA3 genes which are differentially expressed during flower and fruit development. Gene 404:10–24
Rijpkema AS, Royaert S, Zethof J, van der Weerden G, Gerats T, Vandenbussche M (2006) Analysis of the Petunia TM6 MADS box gene reveals functional divergence within the DEF/AP3 lineage. Plant Cell 18:1819–1832
Shan H, Su K, Lu W, Kong H, Chen Z, Meng Z (2006) Conservation and divergence of candidate class B genes in Akebia trifoliate (Lardizabalaceae). Dev Genes Evol 216:785–795
Stellari GM, Jaramillo MA, Kramer EM (2004) Evolution of the APETALA3 and PISTILLATA lineages of MADS-box-containing genes in the basal angiosperms. Mol Biol Evol 21:506–519
Su K, Zhao S, Shan H, Kong H, Lu W, Theissen G, Chen Z, Meng Z (2008) The MIK region rather than the C-terminal domain of AP3-like class B floral homeotic proteins determines functional specificity in the development and evolution of petals. New Phytol 178:544–558
Sui SZ, Li MY, Jiang A, Wang Y, Qin H, Pi W, Ren C (2007) Construction of Chimonanthus praecox flower cDNA library and expression analysis of high-abundance cDNA of lipid transfer Protein (LTP). Scientia Agricultura Sinica 40:644–664 (in Chinese)
Theissen G (2001) Development of floral organ identity: stories from the MADS house. Curr Opin Plant Biol 4:75–85
Theissen G, Saedler H (2001) Floral quartets. Nature 409:469–471
Theissen G, Becker A, Winter KU, Munster T, Kirchner C, Saedler H (2002) How the land plants learned their floral ABCs: the role of MADS-box genes in the evolutionary origin of flowers. In: Cronk QCB, Bateson RM, Hawkins JA (eds) Developmental genetics and plant evolution. Taylor & Francis, London, pp 173–205
Vandenbussche M, Theissen G, Van de Peer Y, Gerats T (2003a) Structural diversification and neo-functionalization during floral MADS-box gene evolution by C-terminal frameshift mutations. Nucleic Acids Res 31:4401–4409
Vandenbussche M, Zethof J, Souer E, Koes R, Tornielli GB, Pezzotti M, Ferrario S, Angenent GC, Gerats T (2003b) Toward the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C, and D floral organ identity functions require SEPALLATA-like MADS box genes in petunia. Plant Cell 15:2680–2693
Whipple CJ, Ciceri P, Padilla CM, Ambrose BA, Bandong SL, Schmidt RJ (2004) Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development 131:6083–6091
Wu CL, Chen WY, Du QP (1999) Studies on the branch and bud characteristics of the wintersweet. Acta Horticulturae Sinica 26:37–42, in Chinese
Wu CL, Hu NZ (1995) Studies on the flower form and blooming characteristics of the wintersweet. Acta Horticulturae Sinica 22:277–282, in Chinese
Xiao H, Wang Y, Liu D, Wang W, Li X, Zhao X, Xu J, Zhai W, Zhu L (2003) Functional analysis of the rice AP3 homologue OsMADS16 by RNA interference. Plant Mol Biol 52:957–966
Zhang RH, Liu HE (1998) Wax shrubs in world (Calycanthaceae). China Science and Technology, Beijing, pp 1–40, in Chinese
Acknowledgments
The authors are grateful to the Arabidopsis Biological Resource Center at the Ohio State University for providing ap3 mutant. The authors thank Dr. Li Song of the Shanghai Institute of Plant Physiology and Ecology for help in microscopic observation and Dr. Fang Duan of York University, Canada for valuable suggestions and critical reading of the manuscript. The authors would also like to thank anonymous reviewers for their valuable comments on the manuscript. This work was partly supported by a grant to Ke Duan from the National Natural Science Foundation of China (no. 30800081).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by K. Schneitz
Qiong Zhang and Bei-Guo Wang contributed equally to this work.
The nucleotide sequence reported in this paper has been submitted to GenBank under the accession number EF059915 (CpAP3).
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
Expression analysis of CpAP3 in wild-type and transgenic petunia (DOC 42 kb)
Rights and permissions
About this article
Cite this article
Zhang, Q., Wang, BG., Duan, K. et al. The paleoAP3-type gene CpAP3, an ancestral B-class gene from the basal angiosperm Chimonanthus praecox, can affect stamen and petal development in higher eudicots. Dev Genes Evol 221, 83–93 (2011). https://doi.org/10.1007/s00427-011-0361-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-011-0361-9