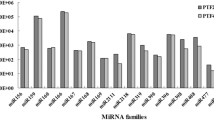
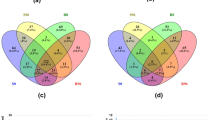
Abstract
Main conclusion
In-depth comparative degradome analysis of two domesticated grape cultivars with diverse secondary metabolite accumulation reveals differential miRNA-mediated targeting.
Abstract
Small (s)RNAs such as micro(mi)RNAs and secondary small interfering (si) often work as negative switches of gene expression. In plants, it is well known that miRNAs target and cleave mRNAs that have high sequence complementarity. However, it is not known if there are variations in miRNA-mediated targeting between subspecies and cultivars that have been subjected to vast genetic modifications through breeding and other selections. Here, we have used PAREsnip2 tool for analysis of degradome datasets derived from two contrasting domesticated grape cultivars having varied fruit color, habit and leaf shape. We identified several interesting variations in sRNA targeting using degradome and 5’RACE analysis between two contrasting grape cultivars that was further correlated using RNA-seq analysis. Several of the differences we identified are associated with secondary metabolic pathways. We propose possible means by which sRNAs might contribute to diversity in secondary metabolites and other development pathways between two domesticated cultivars of grapes.







Similar content being viewed by others
Data availability
sRNA sequencing data for BB and DK grapes are available in Gene Expression Omnibus (GEO) series GSE107907, from GSM2883166 to GSM2883177 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE107907). RNA sequencing data are available in GEO series GSE107905, from GSM2883154 to GSM2883165 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE107905). Degradome data are available in GSE118701 series (from GSM3336816 to GSM333619) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118701). The datasets used for analysis in our current study were from our previous study (Tirumalai et al. 2019).
Abbreviations
- BB:
-
Grape cv Bangalore Blue
- DK:
-
Grape cv Dilkhush
- PhasiRNAs:
-
Phased siRNAs
- RACE:
-
Rapid amplification of cDNA ends
- TasiRNAs:
-
Transacting siRNAs
References
Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ (2008) Endogenous siRNA and miRNA targets Identified by sequencing of the Arabidopsis degradome. Curr Biol 18:758–762. https://doi.org/10.1016/j.cub.2008.04.042
Addo-Quaye C, Miller W, Axtell MJ (2009) CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25:130–131. https://doi.org/10.1093/bioinformatics/btn604
Ahmed W, Li R, Xia Y et al (2020) Comparative analysis of miRNA expression profiles between heat-tolerant and heat-sensitive genotypes of flowering Chinese cabbage under heat stress using high-throughput sequencing. Genes (basel) 11:264. https://doi.org/10.3390/genes11030264
Akbergenov R, Si-Ammour A, Blevins T et al (2006) Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res 34:462–471. https://doi.org/10.1093/nar/gkj447
Albert NW, Davies KM, Schwinn KE (2014a) Repression—the dark side of anthocyanin regulation? Acta Hortic 1048:129–136
Albert NW, Davies KM, Schwinn KE (2014b) Gene regulation networks generate diverse pigmentation patterns in plants. Plant Signal Behav 9:1–3. https://doi.org/10.4161/psb.29526
Ali K, Maltese F, Choi YH, Verpoorte R (2010) Metabolic constituents of grapevine and grape-derived products. Phytochem Rev 9:357–378
Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121:207–221. https://doi.org/10.1016/j.cell.2005.04.004
Axtell MJ, Jan C, Rajagopalan R, Bartel DP (2006) A two-hit trigger for siRNA biogenesis in plants. Cell 127:565–577. https://doi.org/10.1016/j.cell.2006.09.032
Barrera-Figueroa BE, Gao L, Wu Z et al (2012) High throughput sequencing reveals novel and abiotic stress-regulated microRNAs in the inflorescences of rice. BMC Plant Biol 12:132. https://doi.org/10.1186/1471-2229-12-132
Bartel DP (2009) MicroRNAs: Target recognition and regulatory functions. Cell 136:215–233
Baulcombe D (2004) RNA silencing in plants. Nature 431:356–363
Belli Kullan J, Lopes Paim Pinto D, Bertolini E et al (2015) miRVine: a microRNA expression atlas of grapevine based on small RNA sequencing. BMC Genomics 16:393. https://doi.org/10.1186/s12864-015-1610-5
Bonar N, Liney M, Zhang R et al (2018) Potato miR828 is associated with purple tuber skin and flesh color. Front Plant Sci 9:1742. https://doi.org/10.3389/fpls.2018.01742
Bonnet E, He Y, Billiau K, Van de Peer Y (2010) TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 26:1566–1568. https://doi.org/10.1093/bioinformatics/btq233
Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M et al (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 80(320):1185–1190. https://doi.org/10.1126/science.1159151
Cavallini E, Matus JT, Finezzo L et al (2015) The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol 167:1448–1470. https://doi.org/10.1104/pp.114.256172
Čepin U, Gutiérrez-Aguirre I, Balažic L et al (2010) A one-step reverse transcription real-time PCR assay for the detection and quantitation of grapevine fanleaf virus. J Virol Methods 170:47–56. https://doi.org/10.1016/j.jviromet.2010.08.018
Chávez Montes RA, De Fátima R-Cárdenas F, De Paoli E et al (2014) (2014) Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat Commun 51(5):1–15. https://doi.org/10.1038/ncomms4722
Chen X (2005) microRNA biogenesis and function in plants. FEBS Lett 579:5923–5931
Chen H-M, Chen L-T, Patel K et al (2010) 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA 107:15269–15274. https://doi.org/10.1073/pnas.1001738107
Chen GH, Sun JY, Liu M et al (2014) SPOROCYTELESS is a novel embryophyte-specific transcription repressor that interacts with TPL and TCP proteins in Arabidopsis. J Genet Genomics 41:617–625. https://doi.org/10.1016/j.jgg.2014.08.009
Costantini L, Malacarne G, Lorenzi S et al (2015) New candidate genes for the fine regulation of the colour of grapes. J Exp Bot 66:4427–4440. https://doi.org/10.1093/jxb/erv159
Costantini L, Battilana J, Lamaj F et al (2008) Berry and phenology-related traits in grapevine (Vitis vinifera L.): From quantitative trait loci to underlying genes. BMC Plant Biol 8:38. https://doi.org/10.1186/1471-2229-8-38
Creasey KM, Zhai J, Borges F et al (2014) MiRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508:411–415. https://doi.org/10.1038/nature13069
Cuperus JT, Fahlgren N, Carrington JC (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23:431–442
Czemmel S, Stracke R, Weisshaar B et al (2009) The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol 151:1513–1530. https://doi.org/10.1104/pp.109.142059
Deboo GB, Albertsen MC, Taylor LP (1995) Flavanone 3-hydroxylase transcripts and flavonol accumulation are temporally coordinate in maize anthers. Plant J 7:703–713. https://doi.org/10.1046/j.1365-313X.1995.07050703.x
De-La-Cruz Chacón I, Riley-Saldaña CA, González-Esquinca AR (2013) Secondary metabolites during early development in plants. Phytochem Rev 12:47–64
Duan CG, Wang CH, Guo HS (2012) Application of RNA silencing to plant disease resistance. Silence 3:5. https://doi.org/10.1186/1758-907X-3-5
Dubos C, Stracke R, Grotewold E et al (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581
Facchini PJ (1999) Plant secondary metabolism: out of the evolutionary abyss. Trends Plant Sci 4:382–384
Fahlgren N, Montgomery TA, Howell MD et al (2006) Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol 16:939–944. https://doi.org/10.1016/j.cub.2006.03.065
Fei Q, Xia R, Meyers BC (2013) Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25:2400–2415
Feller A, MacHemer K, Braun EL, Grotewold E (2011) Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J 66:94–116. https://doi.org/10.1111/j.1365-313X.2010.04459.x
Gregory BD, O’Malley RC, Lister R et al (2008) A Link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell 14:854–866. https://doi.org/10.1016/j.devcel.2008.04.005
Höll J, Vannozzi A, Czemmel S et al (2013) The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell 25:4135–4149. https://doi.org/10.1105/tpc.113.117127
Kerwin RE, Jimenez-Gomez JM, Fulop D et al (2011) Network quantitative trait loci mapping of circadian clock outputs identifies metabolic pathway-to-clock linkages in Arabidopsis. Plant Cell 23:471–485. https://doi.org/10.1105/TPC.110.082065
Khraiwesh B, Arif MA, Seumel GI et al (2010) Transcriptional control of gene expression by microRNAs. Cell 140:111–122. https://doi.org/10.1016/j.cell.2009.12.023
Kim Y, Kim K, Kim Y et al (2014) Cloning and characterization of a flavonol synthase gene from Scutellaria baicalensis. Sci World J. https://doi.org/10.1155/2014/980740
Kobayashi S, Goto-Yamamoto N, Hirochika H (2004) Retrotransposon-induced mutations in grape skin color. Science 304(5673):982. https://doi.org/10.1126/science.1095011
Kranz HD, Denekamp M, Greco R et al (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J 16:263–276. https://doi.org/10.1046/j.1365-313X.1998.00278.x
Levadoux L (1956) Les populations sauvages et cultivées deVitis vinifera L. Annales De L’amélioration Des Plantes 1:59–116
Liu Y, Lin-Wang K, Espley RV et al (2016) Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J Exp Bot 67:2159–2176. https://doi.org/10.1093/jxb/erw014
Llave C, Xie Z, Kasschau KD, Carrington JC (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053–2056. https://doi.org/10.1126/SCIENCE.1076311/SUPPL_FILE/LLAVE1076311.SUP.PDF
Loris EA, Panjikar S, Ruppert M et al (2007) Structure-based engineering of strictosidine synthase: auxiliary for alkaloid libraries. Chem Biol 14:979–985. https://doi.org/10.1016/j.chembiol.2007.08.009
Luo QJ, Mittal A, Jia F, Rock CD (2012) An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol Biol 80:117–129. https://doi.org/10.1007/s11103-011-9778-9
Ma X, Panjikar S, Koepke J et al (2006) The structure of Rauvolfia serpentina strictosidine synthase is a novel six-bladed β-propeller fold in plant proteins. Plant Cell 18:907–920. https://doi.org/10.1105/tpc.105.038018
Matus JT, Cavallini E, Loyola R et al (2017) A group of grapevine MYBA transcription factors located in chromosome 14 control anthocyanin synthesis in vegetative organs with different specificities compared with the berry color locus. Plant J 91:220–236. https://doi.org/10.1111/tpj.13558
Mi H, Muruganujan A, Thomas PD (2013) PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 41:D377-386. https://doi.org/10.1093/nar/gks1118
Narjala A, Nair A, Tirumalai V et al (2020) A conserved sequence signature is essential for robust plant miRNA biogenesis. Nucleic Acids Res 48:3103–3118. https://doi.org/10.1093/nar/gkaa077
Pantaleo V, Szittya G, Moxon S et al (2010) Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J 62:960–976. https://doi.org/10.1111/j.1365-313X.2010.04208.x
Rock CD (2013) Trans-acting small interfering RNA4: Key to nutraceutical synthesis in grape development? Trends Plant Sci 18:601–610
Romeis T, Ludwig AA, Martin R, Jones JDG (2001) Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J 20:5556–5567. https://doi.org/10.1093/emboj/20.20.5556
Saijo Y, Hata S, Kyozuka J et al (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23:319–327. https://doi.org/10.1046/j.1365-313X.2000.00787.x
Schilmiller AL, Stout J, Weng JK et al (2009) Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J 60:771–782. https://doi.org/10.1111/j.1365-313X.2009.03996.x
Seo JK, Wu J, Lii Y et al (2013) Contribution of small RNA pathway components in plant immunity. Mol Plant-Microbe Interact 26:617–625
Sharma D, Tiwari M, Pandey A et al (2016) MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development. Plant Physiol 171:944–959. https://doi.org/10.1104/pp.15.01831
Shivaprasad PV, Chen HM, Patel K et al (2012) A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24:859–874. https://doi.org/10.1105/tpc.111.095380
Spoor W (2001) Zohary D, Hopf M (2000) Domestication of plants in the Old World, 3rd edn Oxford University Press. Book Review in Ann Bot, New York. https://doi.org/10.1006/anbo.2001.1505
Stöckigt J, Barleben L, Panjikar S, Loris EA (2008) 3D-Structure and function of strictosidine synthase—the key enzyme of monoterpenoid indole alkaloid biosynthesis. Plant Physiol Biochem 46:340–355
Stocks MB, Moxon S, Mapleson D et al (2012) The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics 28:2059–2061. https://doi.org/10.1093/bioinformatics/bts311
Sunitha S, Loyola R, Alcalde JA et al (2018) The role of UV-B light on small RNA activity during grapevine berry development. BioRxiv. https://doi.org/10.1101/375998
Sunkar R, Chinnusamy V, Zhu J, Zhu JK (2007) Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci 12:301–309
Swetha C, Basu D, Pachamuthu K et al (2018) Major domestication-related phenotypes in indica rice are due to loss of miRNA-mediated laccase silencing. Plant Cell 30:2649–2662. https://doi.org/10.1105/tpc.18.00472
Swetha C, Narjala A, Pandit A et al (2022) Degradome comparison between wild and cultivated rice identifies differential targeting by miRNAs. BMC Genomics 23:53. https://doi.org/10.1186/S12864-021-08288-5
Tähtiharju S, Sangwan V, Monroy AF et al (1997) The induction of kin genes in cold-acclimating Arabidopsis thaliana. Evidence of a role for calcium. Planta 203:442–447. https://doi.org/10.1007/s004250050212
Tao Q, Guo D, Wei B et al (2013) The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25:421–437. https://doi.org/10.1105/tpc.113.109223
Taylor RS, Tarver JE, Hiscock SJ, Donoghue PCJ (2014) Evolutionary history of plant microRNAs. Trends Plant Sci 19:175–182
Terrier N, Torregrosa L, Ageorges A et al (2009) Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol 149:1028–1041. https://doi.org/10.1104/pp.108.131862
Thody J, Folkes L, Medina-Calzada Z et al (2018) PAREsnip2: a tool for high-throughput prediction of small RNA targets from degradome sequencing data using configurable targeting rules. Nucleic Acids Res 46:8730–8739. https://doi.org/10.1093/nar/gky609
Tirumalai V, Swetha C, Nair A et al (2019) MiR828 and miR858 regulate VvMYB114 to promote anthocyanin and flavonol accumulation in grapes. J Exp Bot 70:4775–4791. https://doi.org/10.1093/jxb/erz264
Tirumalai V, Prasad M, Shivaprasad PV (2020) Rna blot analysis for the detection and quantification of plant micrornas. J vis Exp 2020:1–8. https://doi.org/10.3791/61394
Wang Y, Zhang WZ, Song LF et al (2008) Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol 148:1201–1211. https://doi.org/10.1104/pp.108.126375
Wang Y, Itaya A, Zhong X et al (2011) Function and evolution of a microRNA that regulates a caspi2+-ATPase and triggers the formation of phased small interfering RNAs in tomato reproductive growth. Plant Cell 23:3185–3203. https://doi.org/10.1105/tpc.111.088013
Xia R, Zhu H, An Y, qiang, et al (2012) Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol 13:r47. https://doi.org/10.1186/gb-2012-13-6-r47
Xia R, Meyers BC, Liu Z et al (2013) MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA biogenesis in eudicots. Plant Cell 25:1555–1572. https://doi.org/10.1105/tpc.113.110957
Xia R, Ye S, Liu Z et al (2015) Novel and recently evolved microRNA clusters regulate expansive F-BOX gene networks through phased small interfering RNAs in wild diploid strawberry. Plant Physiol 169:594–610. https://doi.org/10.1104/pp.15.00253
Yan X, Qiao H, Zhang X et al (2017) Analysis of the grape (Vitis vinifera L.) thaumatin-like protein (TLP) gene family and demonstration that TLP29 contributes to disease resistance. Sci Rep 7(1):4269. https://doi.org/10.1038/s41598-017-04105-w
Yao G, Ming M, Allan AC et al (2017) Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J 92:437–451. https://doi.org/10.1111/TPJ.13666
Yu XC, Li MJ, Gao GF et al (2006) Abscisic acid stimulates a calcium-dependent protein kinase in grape berry. Plant Physiol 140:558–579. https://doi.org/10.1104/pp.105.074971
Zhai J, Arikit S, Simon SA et al (2014) Rapid construction of parallel analysis of RNA end (PARE) libraries for Illumina sequencing. Methods 67:84–90. https://doi.org/10.1016/j.ymeth.2013.06.025
Zhang C, Li G, Wang J, Fang J (2012) Identification of trans-acting siRNAs and their regulatory cascades in grapevine. Bioinformatics 28:2561–2568. https://doi.org/10.1093/bioinformatics/bts500
Zheng Y, Wang S, Sunkar R (2014) Genome-wide discovery and analysis of phased small interfering RNAs in Chinese sacred lotus. PLoS ONE 9:e113790. https://doi.org/10.1371/journal.pone.0113790
Acknowledgements
The authors acknowledge Genotypic Technologies, Bangalore for sRNA and RNA-seq. Thanks to Dr. Awadhesh Pandit, NGS facility, NCBS for performing the degradome sequencing.
Funding
PVS lab is supported by NCBS-TIFR core funding and grants (BT/PR12394/AGIII/103/891/2014; BT/IN/Swiss/47/JGK/2018–19; BT/PR25767/GET/ 119/151/2017) from Department of Biotechnology, Government of India. We also acknowledge support of the Department of Atomic Energy, Government of India, under Project Identification No. RTI 4006 (1303/3/2019/R&D-II/DAE/4749 dated 16.7.2020). AN and SC acknowledge a research fellowship from DBT.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tirumalai, V., Narjala, A., Swetha, C. et al. Cultivar-specific miRNA-mediated RNA silencing in grapes. Planta 256, 17 (2022). https://doi.org/10.1007/s00425-022-03934-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-022-03934-y