Abstract
Main conclusion
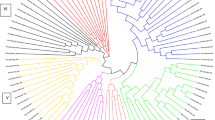
Analysis of 180 accessions of Miscanthus using a DArT platform revealed high diversity. The phylogenetic analysis revealed that M. × giganteus accessions fall into two genetically distinct groups.
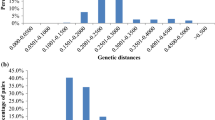
Miscanthus is a genus of perennial rhizomatous grasses that has emerged in last 20 years as a feedstock for bioenergy and biofuel production. Currently, the most widely used accession for bioenergy purposes is Miscanthus × giganteus, a sterile triploid hybrid between Miscanthus sinensis and Miscanthus sacchariflorus. However, previous reports have shown that genetic diversity of Miscanthus × giganteus is limited. Here, we report development of Diversity Arrays Technology platform for the analysis of genetic structure of a Miscanthus collection of 180 accessions. A total of 906 markers were obtained of which around 25.5 % exhibited polymorphism information content value in the range of 0.40 and 0.50 and are considered particularly informative. Newly developed marker system will serve as an additional resource to assist crop improvement, germplasm preservation and genetic studies. Three types of analysis indicated that 180 accessions from the collection were well differentiated and presented high diversity. Interestingly, the analysis revealed that there are two separate groups of plants, significantly differing in genetic diversity, that are commercially available as M. × giganteus. We suggest that one of these groups is most likely mutants or somaclonal variants of original M. × giganteus. The other group is recent hybrids of Miscanthus of higher genetic diversity. This study indicates that the diversity of commercially available M. × giganteus is higher than commonly assumed. Development of the new marker system can significantly assist breeding of new commercial cultivars of Miscanthus for bioenergy use.




Similar content being viewed by others
Abbreviations
- AMOVA:
-
Analysis of molecular variance
- DArT:
-
Diversity arrays technology
- PCoA:
-
Principal coordinate analysis
- PIC:
-
Polymorphism information content
- RAD-seq:
-
Restriction site associated DNA sequencing
- UPGMA:
-
Unweighted pair group method with algorithmic mean
References
Atienza S, Satovic Z, Petersen K, Dolstra O, Martin A (2002) Preliminary genetic linkage map of Miscanthus sinensis with RAPD markers. Theor Appl Genet 105:946–952
Castillo A, Ramírez MC, Martín AC, Kilian A, Martín A, Atienza SG (2013) High-throughput genotyping of wheat-barley amphiploids utilising diversity array technology (DArT). BMC Plant Biol 13:87
Chiang YC, Chou CH, Lee PR, Chiang TY (2001) Detection of leaf-associated fungi based on PCR and nucleotide sequence of the ribosomal internal transcribed spacer (ITS) in Miscanthus. Bot Bull Acad Sinica 42:39–44
Chouvarine P, Cooksey AM, McCarthy FM, Ray DA, Baldwin BS, Burgess SC, Peterson DG (2012) Transcriptome-based differentiation of closely-related Miscanthus lines (transcriptome-based differentiation of Miscanthus). PLoS One 7:e29850
Clifton-Brown JC, Lewandowski I, Andersson B, Basch G, Christian DG, Kjeldsen JB, Jorgensen U, Mortensen JV, Riche AB, Schwarz KU, Tayebi K, Teixeira F (2001) Performance of 15 Miscanthus genotypes at five sites in Europe. Agron J 93:1013–1019
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297–302
Dominguez-Garcia MC, Belaj A, De la Rosa R, Satovic Z, Heller-Uszynska K, Kilian A, Martin A, Atienza SG (2012) Development of DArT markers in olive (Olea europaea L.) and usefulness in variability studies and genome mapping. Sci Hortic 136:50–60
Earl DA (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Głowacka K, Clark LV, Adhikari S, Peng J, Ryan Stewart J, Nishiwaki A, Yamada T, Jørgensen U, Hodkinson TR, Gifford J (2015) Genetic variation in Miscanthus × giganteus and the importance of estimating genetic distance thresholds for differentiating clones. GCB Bioenergy 7:386–404
Greef J, Deuter M (1993) Syntaxonomy of Miscanthus × giganteus Greef et Deu. Angew Bot 67:87–90
Greef J, Deuter M, Jung C, Schondelmaier J (1997) Genetic diversity of European Miscanthus species revealed by AFLP fingerprinting. Genet Resour Crop Ev 44:185–195
Grzebelus D, Iorizzo M, Senalik D, Ellison S, Cavagnaro P, Macko-Podgorni A, Heller-Uszynska K, Kilian A, Nothnagel T, Allender C (2014) Diversity, genetic mapping, and signatures of domestication in the carrot (Daucus carota L.) genome, as revealed by Diversity Arrays Technology (DArT) markers. Mol Breeding 33:625–637
Hauser LA, Crovello TJ (1982) Numerical analysis of generic relationships in Thelypodieae (Brassicaceae). Syst Bot 7:249–268
Heaton EA, Dohleman FG, Long SP (2008a) Meeting US biofuel goals with less land: the potential of Miscanthus. Glob Change Biol 14:2000–2014
Heaton EA, Flavell RB, Mascia PN, Thomas SR, Dohleman FG, Long SP (2008b) Herbaceous energy crop development: recent progress and future prospects. Curr Opin Biotech 19:202–209
Heller-Uszynska K, Uszynski G, Huttner E, Evers M, Carlig J, Caig V, Aitken K, Jackson P, Piperidis G, Cox M, Gilmour R, D’Hont A, Butterfield M, Glaszmann JC, Kilian A (2011) Diversity Arrays Technology effectively reveals DNA polymorphism in a large and complex genome of sugarcane. Mol Breed 28:37–55
Hernández P, Dorado G, Laurie D, Martin A, Snape J (2001) Microsatellites and RFLP probes from maize are efficient sources of molecular markers for the biomass energy crop Miscanthus. Theor Appl Genet 102:616–622
Hodkinson TR, Chase MW, Lledó DM, Salamin N, Renvoize SA (2002) Phylogenetics of Miscanthus, Saccharum and related genera (Saccharinae, Andropogoneae, Poaceae) based on DNA sequences from ITS nuclear ribosomal DNA and plastid trnL intron and trnL-F intergenic spacers. J Plant Res 115:381–392
Jaccoud D, Peng K, Feinstein D, Kilian A (2001) Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res 29:e25–e25
James KE, Schneider H, Ansell SW, Evers M, Robba L, Uszynski G, Pedersen N, Newton AE, Russell SJ, Vogel JC (2008) Diversity arrays technology (DArT) for pan-genomic evolutionary studies of non-model organisms. PLoS One 3:e1682
Jeżowski S (2008) Yield traits of six clones of Miscanthus in the first 3 years following planting in Poland. Ind Crops Prod 27:65–68
Jeżowski S, Glowacka K, Kaczmarek Z (2009) Wstępna ocena głównych parametrów wymiany gazowej związanych z fotosyntezą w odniesieniu do plonowania traw energetycznych z rodzaju Miscanthus w pierwszym roku uprawy. Acta Agrophysica 14:73–81
Jeżowski S, Głowacka K, Kaczmarek Z (2011) Variation on biomass yield and morphological traits of energy grasses from the genus Miscanthus during the first years of crop establishment. Biomass Bioenergy 35:814–821
Jones MB, Finnan J, Hodkinson TR (2015) Morphological and physiological traits for higher biomass production in perennial rhizomatous grasses grown on marginal land. GCB Bioenergy 7:375–385
Kilian A, Wenzl P, Huttner E, Carling J, Xia L, Blois H, Caig V, Heller-Uszynska K, Jaccoud D, Hopper C, Aschenbrenner-Kilian M, Evers M, Peng K, Cayla C, Hok P, Uszynski G (2012) Diversity Arrays Technology: a generic genome profiling technology on open platforms. In: Pompanon F, Bonin A (eds) Data production and analysis in population genomics. Humana Press, New York, pp 67–89
Lee NY (1964a) Taxonomic studies on the genus Miscanthus: anatomical patterns of leaves. Bot Mag Tokyo 77:122–130
Lee NY (1964b) Taxonomic studies on the genus Miscanthus: relationships among the section, subsection and species, part 1. J Jpn Bot 39:196–205
Lee NY (1964c) Taxonomic studies on the genus Miscanthus: relationships among the section, subsection and species, part 2. J Jpn Bot 39:257–268
Lee NY (1964d) Taxonomic studies on the genus Miscanthus: relationships among the section, subsection and species, part 3. J Jpn Bot 39:289–298
Lewandowski I, Clifton-Brown JC, Andersson B, Basch G, Christian DG, Jorgensen U, Jones MB, Riche AB, Schwarz KU, Tayebi K, Teixeira F (2003) Environment and harvest time affects the combustion qualities of Miscanthus genotypes. Agron J 95:1274–1280
Mace ES, Xia L, Jordan DR, Halloran K, Parh DK, Huttner E, Wenzl P, Kilian A (2008) DArT markers: diversity analyses and mapping in Sorghum bicolor. BMC Genom 9:26
Matlaga DP, Davis AS (2013) Minimizing invasive potential of Miscanthus × giganteus grown for bioenergy: identifying demographic thresholds for population growth and spread. J App Ecol 50:479–487
Mos M, Banks SW, Nowakowski DJ, Robson PRH, Bridgwater AV, Donnison IS (2013) Impact of Miscanthus × giganteus senescence times on fast pyrolysis bio-oil quality. Bioresour Technol 129:335–342
Oliver R, Jellen E, Ladizinsky G, Korol A, Kilian A, Beard J, Dumlupinar Z, Wisniewski-Morehead N, Svedin E, Coon M (2011) New Diversity Arrays Technology (DArT) markers for tetraploid oat (Avena magna Murphy et Terrell) provide the first complete oat linkage map and markers linked to domestication genes from hexaploid A. sativa L. Theor Appl Genet 123:1159–1171
Ovesná J, Kučera L, Vaculová K, Milotová J, Snape J, Wenzl P, Huttner E, Kilian A, Martelli G, Milella L (2013) Analysis of the genetic structure of a barley collection using DNA Diversity Array Technology (DArT). Plant Mol Biol Rep 31:280–288
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Quinn LD, Allen DJ, Stewart JR (2010) Invasiveness potential of Miscanthus sinensis: implications for bioenergy production in the United States. GCB Bioenergy 2:310–320
Raman H, Stodart B, Cavanagh C, Mackay M, Morell M, Milgate A, Martin P (2010) Molecular diversity and genetic structure of modern and traditional landrace cultivars of wheat (Triticum aestivum L.). Crop Pasture Sci 61:222–229
Rohlf F (1997) NTSYS-pc 2.1. Numerical taxonomy and multivariate analysis system Exeter Software. Exeter publishing Setauket, NY
Sang T, Zhu W (2011) China’s bioenergy potential. GCB Bioenergy 3:79–90
Sharma M, Nagavardhini A, Thudi M, Ghosh R, Pande S, Varshney RK (2014) Development of DArT markers and assessment of diversity in Fusarium oxysporum f. sp. ciceris, wilt pathogen of chickpea (Cicer arietinum L.). BMC Genom 15:454
Simko I, Eujayl I, van Hintum TJ (2012) Empirical evaluation of DArT, SNP, and SSR marker-systems for genotyping, clustering, and assigning sugar beet hybrid varieties into populations. Plant Sci 184:54–62
Von Mark VC, Kilian A, Dierig DA (2013) Development of DArT marker platforms and genetic diversity assessment of the US collection of the new oilseed crop Lesquerella and related species. PLoS One 8:e64062
Wenzl P, Carling J, Kudrna D, Jaccoud D, Huttner E, Kleinhofs A, Kilian A (2004) Diversity arrays technology (DArT) for whole-genome profiling of barley. Proc Natl Acad Sci USA 101:9915–9920
Xia L, Peng K, Yang S, Wenzl P, De Vicente MC, Fregene M, Kilian A (2005) DArT for high-throughput genotyping of cassava (Manihot esculenta) and its wild relatives. Theor Appl Genet 110:1092–1098
Zhao H, Yu JY, You FM, Luo MC, Peng JH (2011) Transferability of microsatellite markers from Brachypodium distachyon to Miscanthus sinensis, a potential biomass crop. J Integr Plant Biol 53:232–245
Acknowledgments
This study was mainly funded by Energene sp. z o.o., and partially supported by Peking University Shenzhen Graduate School start up grant 0068 to MD and a Key Project of Shenzhen Emerging Industries (No. JC201104210118A). MM holds executive position and has financial interest in Energene.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
J. Tang and M. Daroch were contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tang, J., Daroch, M., Kilian, A. et al. DArT-based characterisation of genetic diversity in a Miscanthus collection from Poland. Planta 242, 985–996 (2015). https://doi.org/10.1007/s00425-015-2335-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-015-2335-z