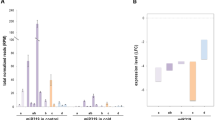
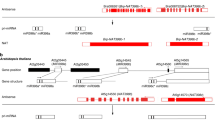
Abstract
Main conclusion
Our work strongly suggests that microRNA858 regulates anthocyanin biosynthesis in tomato by modulating the expression of two R2R3 MYB transcription factors, underscoring the importance of microRNAs in the gene regulatory network controlling specialized metabolism in plants.
The biological functions of microRNA858 (miR858), a recently identified small RNA, are not well understood. Here, we identified miR858 as a negative regulator of anthocyanin biosynthesis in tomato (Solanum lycopersicum). RNA ligase-mediated 5′RACE cleavage assay showed that miR858 mediates the cleavage of SlMYB7-like and SlMYB48-like transcripts in tomato. Expression analysis revealed an inverse correlation between the accumulation of miR858 and its target SlMYB7-like mRNA, in different tissues of tomato. Subsequently, a small tandem target mimic construct for blocking miR858 (STTM858) was generated and transformed into tomato. The majority of endogenous miR858 was blocked in STTM858 over-expressing tomato plants, whereas SlMYB7-like transcripts increased significantly. Concomitantly, upregulated expression was detected for several anthocyanin biosynthetic genes, including PAL, CHS, DFR, ANS and 3GT. As a result, anthocyanins were highly accumulated in young seedlings, leaves, stems and leaf buds of the transgenic plants under normal growth conditions. In addition, over-expression of STTM858 in tomato activated another MYB transcription factor, SlMYB48, implicating the possible involvement of SlMYB48 in anthocyanin biosynthesis.





Similar content being viewed by others
Abbreviations
- ANS:
-
Anthocyanidin synthase
- CHS:
-
Chalcone synthase
- DFR:
-
Dihydroflavonol reductase
- miRNA:
-
MicroRNA
- PAL:
-
Phenylalanine ammonialyase
- pri-miRNA:
-
Primary miRNA
- qRT-PCR:
-
Quantitative real-time reverse transcription PCR
- STTM:
-
Small tandem target mimic
- 3GT:
-
Flavonol-3-glucosyltransferase
References
Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18:758–762
Ballester AR, Molthoff J, de Vos R, Hekkert B, Orzaez D, Fernandez-Moreno JP, Tripodi P, Grandillo S, Martin C, Heldens J, Ykema M, Granell A, Bovy A (2010) Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol 152:71–84
Baulcombe D (2004) RNA silencing in plants. Nature 431:356–363
Buer CS, Imin N, Djordjevic MA (2010) Flavonoids: new roles for old molecules. J Integr Plant Biol 52:98–111
Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, Schijlen EG, Hall RD, Bovy AG, Luo J, Martin C (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26:1301–1308
Castellarin SD, Pfeiffer A, Sivilotti P, Degan M, Peterlunger E, Dig G (2007) Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine under seasonal water deficit. Plant, Cell Environ 30:1381–1399
Christie P, Alfenito M, Walbot V (1994) Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways: enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta 194:541–549
Distelfeld A, Pearce SP, Avni R, Scherer B, Uauy C, Piston F, Slade A, Zhao R, Dubcovsky J (2012) Divergent functions of orthologous NAC transcription factors in wheat and rice. Plant Mol Biol 78:515–524
Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55:940–953
Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2:e219
Fahlgren N, Jogdeo S, Kasschau KD, Sullivan CM, Chapman EJ, Laubinger S, Smith LM, Dasenko M, Givan SA, Weigel D, Carrington JC (2010) MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 22:1074–1089
Fornale S, Lopez E, Salazar-Henao JE, Fernandez-Nohales P, Rigau J, Caparros-Ruiz D (2014) AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant Cell Physiol 55:507–516
Garcia-Mas J, Benjak A, Sanseverino W, Bourgeois M, Mir G, Gonzalez VM, Henaff E, Camara F, Cozzuto L, Lowy E, Alioto T, Capella-Gutierrez S, Blanca J, Canizares J, Ziarsolo P, Gonzalez-Ibeas D, Rodriguez-Moreno L, Droege M, Du L, Alvarez-Tejado M, Lorente-Galdos B, Mele M, Yang L, Weng Y, Navarro A, Marques-Bonet T, Aranda MA, Nuez F, Pico B, Gabaldon T, Roma G, Guigo R, Casacuberta JM, Arus P, Puigdomenech P (2012) The genome of melon (Cucumis melo L.). Proc Natl Acad Sci USA 109:11872–11877
Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23:1512–1522
Guan X, Pang M, Nah G, Shi X, Ye W, Stelly DM, Chen ZJ (2014) miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat Commun 5:3050
He J, Silva AM, Mateus N, de Freitas V (2011) Oxidative formation and structural characterisation of new alpha-pyranone (lactone) compounds of non-oxonium nature originated from fruit anthocyanins. Food Chem 127:984–992
Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ (2009) Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151:2120–2132
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Li YY, Mao K, Zhao C, Zhao XY, Zhang HL, Shu HR, Hao YJ (2012) MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol 160:1011–1022
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25:402–408
Lorenc-Kukula K, Jafra S, Oszmianski J, Szopa J (2005) Ectopic expression of anthocyanin 5-o-glucosyltransferase in potato tuber causes increased resistance to bacteria. J Agric Food Chem 53:272–281
Luo QJ, Mittal A, Jia F, Rock CD (2012) An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol Biol 80:117–129
Maier A, Schrader A, Kokkelink L, Falke C, Welter B, Iniesto E, Rubio V, Uhrig JF, Hulskamp M, Hoecker U (2013) Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J 74:638–651
Mathews H, Clendennen SK, Caldwell CG, Liu XL, Connors K, Matheis N, Schuster DK, Menasco DJ, Wagoner W, Lightner J, Wagner DR (2003) Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15:1689–1703
Matsui K, Umemura Y, Ohme-Takagi M (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55:954–967
Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ (2009) RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol 21:367–376
Mehrtens F, Kranz H, Bednarek P, Weisshaar B (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138:1083–1096
Meiers S, Kemeny M, Weyand U, Gastpar R, von Angerer E, Marko D (2001) The anthocyanidins cyanidin and delphinidin are potent inhibitors of the epidermal growth-factor receptor. J Agric Food Chem 49:958–962
Mooney M, Desnos T, Harrison K, Jones J, Carpenter R, Coen E (1995) Altered regulation of tomato and tobacco pigmentation genes caused by the delila gene of Antirrhinum. Plant J 7:333–339
Morishita T, Kojima Y, Maruta T, Nishizawa-Yokoi A, Yabuta Y, Shigeoka S (2009) Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol 50:2210–2222
Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T (2008) Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res 18:1602–1609
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol Plant 15:473–497
Oh S, Park S, Han KH (2003) Transcriptional regulation of secondary growth in Arabidopsis thaliana. J Exp Bot 54:2709–2722
Patra B, Pattanaik S, Yuan L (2013a) Proteolytic degradation of the flavonoid regulators, TRANSPARENT TESTA8 and TRANSPARENT TESTA GLABRA1, in Arabidopsis is mediated by the ubiquitin/26Sproteasome system. Plant Signal Behav. doi:10.4161/psb25901
Patra B, Pattanaik S, Yuan L (2013b) Ubiquitin protein ligase 3 mediates the proteasomal degradation of GLABROUS 3 and ENHANCER OF GLABROUS 3, regulators of trichome development and flavonoid biosynthesis in Arabidopsis. Plant J 74:435–447
Patra B, Schluttenhofer C, Wu Y, Pattanaik S, Yuan L (2013c) Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim Biophys Acta 1829:1236–1247
Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D (2011) The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23:1795–1814
Rajagopalan R, Vaucheret H, Trejo J, Bartel DP (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20:3407–3425
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110:513–520
Sarma AD, Sharma R (1999) Anthocyanin-DNA copigmentation complex: mutual protection against oxidative damage. Phytochemistry 52:1313–1318
Slavikova S, Ufaz S, Avin-Wittenberg T, Levanony H, Galili G (2008) An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J Exp Bot 59:4029–4043
Smith CJ, Watson CF, Bird CR, Ray J, Schuch W, Grierson D (1990) Expression of a truncated tomato polygalacturonase gene inhibits expression of the endogenous gene in transgenic plants. Mol Gen Genet 224:477–481
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4:447–456
Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50:660–677
Tanaka Y, Ohmiya A (2008) Seeing is believing: engineering anthocyanin and carotenoid biosynthetic pathways. Curr Opin Biotechnol 19:190–197
Tang G, Yan J, Gu Y, Qiao M, Fan R, Mao Y, Tang X (2012) Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods 58:118–125
Tominaga-Wada R, Nukumizu Y, Wada T (2013) Tomato (Solanum lycopersicum) homologs of TRIPTYCHON (SlTRY) and GLABRA3 (SlGL3) are involved in anthocyanin accumulation. Plant Signal Behav 8:e24575
Xia R, Zhu H, An YQ, Beers EP, Liu Z (2012) Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol 13:R47
Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 20:176–185
Yang F, Cai J, Yang Y, Liu Z (2013) Overexpression of microRNA828 reduces anthocyanin accumulation in Arabidopsis. Plant Cell Tissue Organ Cult (PCTOC) 115:159–167
Zhu H, Xia R, Zhao B, An YQ, Dardick CD, Callahan AM, Liu Z (2012) Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs. BMC Plant Biol 12:149
Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40:22–34
Zuluaga D, Gonzali S, Loreti E, Pucciariello C, Innocenti E, Guidi L, Alpi A, Perata P (2008) Arabidopsis thaliana MYB75/PAP1 transcription factor induces anthocyanin production in transgenic tomato plants. Funct Plant Biol 35:606–618
Acknowledgments
X. J. is supported by NSFC (31101555), NSF of Shanxi Province (2011021032-2), Program for Leading Young Scientists of Shanxi High Education (2012), Returned Overseas Scholar Research Fund of Shanxi (2011051), Talent Introduction and Development of Shanxi (614191), Science and Technology Innovation Fund of Shanxi Agricultural University (2010021). Research in L. Y. lab is partially supported by a grant from the United States National Science Foundation (Award number 1355438) and the Kentucky Tobacco Research and Development Center.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jia, X., Shen, J., Liu, H. et al. Small tandem target mimic-mediated blockage of microRNA858 induces anthocyanin accumulation in tomato. Planta 242, 283–293 (2015). https://doi.org/10.1007/s00425-015-2305-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-015-2305-5