Abstract
Background
Edaravone was approved as a new treatment for amyotrophic lateral sclerosis (ALS), although there are different opinions on its effectiveness. Magnetic resonance (MRI) measures appear promising as diagnostic and prognostic indicators of disease. However, published studies on MRI using to monitor treatment efficacy in ALS are lacking.
Purpose
The objective of this study was to investigate changes in brain MRI measures in patients treated with edaravone.
Methods
Thirteen ALS patients assuming edaravone (ALS-EDA) underwent MRI at baseline (T0) and after 6 months (T6) to measure cortical thickness (CT) and fractional anisotropy (FA) of white matter (WM) tracts. MRI data of ALS-EDA were compared at T0 with those of 12 control subjects (CS), and at T6 with those of 11 ALS patients assuming only riluzole (ALS-RIL), extracted from our ALS cohort using a propensity-score-matching. A longitudinal MRI analysis was performed in ALS-EDA between T6 and T0.
Results
At T0, ALS-EDA showed a cortical widespread thinning in both hemispheres, particularly in the bilateral precentral gyrus, and a reduction of FA in bilateral corticospinal tracts, in comparison to CS. Thinning in bilateral precentral cortex and significant widespread reduction of FA in several WM tracts were observed in ALS-EDA at T6 compared to T0. At T6, no significant differences in MRI measures of ALS-EDA versus ALS-RIL were found.
Conclusions
Patients treated with edaravone showed progression of damage in the motor cortex and several WM tracts, at a six-month follow-up. Moreover, this study showed no evidence of a difference between edaravone and riluzole.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease causing weakness and wasting of voluntary muscle, associated in about 50% of cases with cognitive impairment [1]. Although the pathogenic mechanism is not yet understood, oxidative stress caused by free radicals seems to be an essential factor involved in motor neuron degeneration and in the progression of the disease [2].
Edaravone is a free-radical scavenger of peroxyl radical that could potentially reduce the postulated oxidative stress in ALS. For this reason, edaravone has been proposed, approved and licensed as the first new treatment for ALS after riluzole. The first phase 3 study did not show significant differences in the ALSFRS-r score between patients receiving edaravone and placebo; however, post hoc analyses identified a subpopulation in which edaravone showed efficacy [3, 4]. To date, there are several reports on the clinical efficacy and safety of edaravone, with promising but discordant results [5,6,7,8]. More recently, no differences in ALS patients treated and not treated with edaravone in terms of disease progression and respiratory function has been reported in a multicenter Italian real-life study [9].
Magnetic resonance imaging (MRI) plays a role in the diagnostic work-up of ALS and currently, its importance is further growing up by the advent of neuroimaging putative biomarkers [10]. Newer MRI techniques appear promising as diagnostic and prognostic indicators of disease. In particular, surface-based cortical thickness measures, voxel-based morphometry for regional grey matter volume variations and diffusion tensor imaging (DTI) metrics of white matter tracts have gained wide consensus [11]. To our knowledge, there are no published studies on MRI measures as an indicator of the effectiveness of drugs in ALS.
The objective of this study was to investigate changes in brain MRI measures in patients treated with edaravone.
Materials and methods
Population
Three groups of subjects were studied: (a) ALS patients taking edaravone in addition to riluzole (13 patients; ALS-EDA); (b) control subjects (12 subjects; CS); (c) ALS patients taking only riluzole (11 patients; ALS-RIL). All subjects included in the study underwent MRI. ALS patients were recruited at the tertiary center of motor neuron diseases of our Department.
Thirteen patients, who met the criteria of the Italian Drug Agency for edaravone administration, were enrolled in the first group (ALS-EDA). Inclusion criteria were clinically “probable” or “definite” ALS according to revised El Escorial Criteria [12], age > 18 years, disease duration less than 2 years, forced vital capacity (FVC) ≥ 80% predicted normal value for gender-height-age in a seated position at the screening visit, a subscore ≥ 2 in all items of ALSFRS-r score and a decrease in the ALSFRS-r score of 1–4 during a 12-week observation period between the screening and the baseline. Exclusion criteria were concomitant significant neurological or neurodegenerative diseases, concomitant significant diseases in other systems or organs, creatine clearance lower than 50 mL/min, pregnant or breastfeeding women, and patients who do not understand or provide informed consent. At inclusion, all patients had to take riluzole 100 mg/day for at least one month. These patients underwent clinical evaluations by a neurologist of the ALS team and MRI investigations at baseline (T0, in 13 patients) and after 6 months (T6, in 11 patients). The following clinical data were collected: site of onset (bulbar or spinal), sex, age, disease duration from symptoms onset, ALSFRS-r, progression rate [13]. Furthermore, each patient has been evaluated for a degree of UMN burden, using Penn Upper Motor Neuron score [14], stratifying the study population using 50% of the total score as cut-off. At follow-up, two patients dropped out, one due to death and one because of inability to perform magnetic resonance imaging for severe orthopnea.
Controls (CS) were age and sex-matched subjects to edaravone group, and consisted of 12 subjects not affected by neurodegenerative diseases, family history of ALS, and with normal brain MRI, that was performed in the diagnostic work-up of their disease.
The third group included 11 definite or probable ALS patients, extracted from a cohort of 40 patients of our Apulia ALS registry, all treated with riluzole (ALS-RIL), who underwent the same MRI protocol of ALS-EDA group. The comparative MRI findings at T6 of ALS-EDA versus ALS-RIL was assessed in propensity score-matched groups. The aforementioned clinical data were collected for ALS-RIL patients.
All ALS patients underwent physiotherapy from the diagnosis of the disease at our tertiary centre; however, the considering the lack of a standardized protocol for this treatment, this latter was not included as variable influencing MRI outcome.
Written informed consent was obtained from each participant according to the Declaration of Helsinki and the study was approved by the Interregional Independent Ethical Committee of “Azienda Ospedaliero- Universitaria” of Bari-Italy.
MRI acquisitions
All participants underwent MRI on a Philips MR system 1.5 T scanner. CS and ALS-RIL groups underwent the same brain MRI protocol once, while ALS-EDA patients twice, at T0 and T6.
Routine T1, T2 weighted sequences and fluid-attenuated inversion recovery (FLAIR) were performed to exclude unrelated abnormalities. 3D-structural MRI was acquired using a T1-weighted MP-RAGE (magnetization-prepared rapid acquisition with gradient echo) sequence (TR/TE/flip angle: 25.00 ms/4.60 ms/30.00 degree; Field Of View [FOV]:240 mm; matrix 256 × 256, voxel size 0.93 × 0.93x1.0 mm3). For Diffusion Tensor Imaging (DTI) analyses, we used a spin echo DTI sequence: 1.75 × 1.75 × 2.5 mm3 acquiring voxel size, reconstructed matrix 128 × 128; 60 slices; TE/TR 77.791/3461 ms; flip angle 90°; a DTI diffusion scheme was used, and a total of 16 diffusion sampling directions were acquired. The b value was 800 s/mm2. The in-plane resolution was 1.75 mm. The slice thickness was 2.5 mm.
Cortical thickness (CT) analysis
FreeSurfer software v.7.1 was employed to assess CT. Processing steps included correction for magnetic field inhomogeneity, alignment to a specific atlas [15], skull removal and segmentation of voxels into grey matter (GM), white matter (WM), and cerebrospinal fluid (CSF). CT was then calculated based on the shortest distance of two surfaces: the interface between GM and WM and the pial surface. A Gaussian filter of 10 mm full width at half maximum was used for smoothing in all analyses.
We employed the longitudinal FreeSurfer pipeline to evaluate 6-month CT changes. The software co-registers the two-time points scan for each subject using a robust and inverse consistent registration algorithm to create an unbiased subject-specific template [16, 17]. Then, several steps in the longitudinal processing stream are initialised from a subject-specific template. This approach has been shown to increase reliability and statistical power [18].
Cerebral WM analysis
The DTI datasets were processed with the FMRIB Software Library v6.0 (FSL) software package. Pre-processing included denoising, removing Gibb’s ringing artefacts, eddy currents, and motion correction. Afterwards, a diffusion tensor model was fitted at each voxel, generating maps of fractional anisotropy (FA), performed with the tract based spatial statistics (TBSS) algorithm, as described elsewhere [19]. Our longitudinal TBSS pipeline follows the steps proposed by Menke et al. [20]. FA maps of each patient in native space were linearly registered into halfway space and averaged. To accomplish that, both images were linearly registered to each other, and then the transformation matrix into halfway space was calculated. This transformation was applied in both images, thus resulting in an average image. Afterwards, we ran the standard TBSS protocol.
Statistical analysis
In descriptive clinical analyses, continuous variables were summarised as mean ± SD as median and range, and categorical variables were expressed as relative frequencies. Mann–Whitney and Fisher’s exact test were assessed for comparisons demographic and clinical features between ALS-EDA at T0 and CS groups, between ALS-EDA at T6 and ALS-RIL groups.
(a) The first step of the study included a cross-sectional analysis of MRI findings in ALS-EDA at T0 versus CS. Furthermore to define the effect of edaravone as addition therapy to riluzole on MRI patterns, ALS-EDA at T6 were compared to ALS-RIL, this latter including patients taking riluzole for the same duration of ALS-EDA. Then, to allow for an unbiased comparison, ALS-RIL were selected from a pool of 40 ALS patients using a propensity score-matched on a one-to-one basis, at the time of MRI concerning riluzole treatment duration. One-to-one matching was performed based on nearest neighbour matching within a calliper of a width of 0.2 standard deviations [21]. Site of onset (bulbar or spinal), sex, age, disease duration from symptoms onset, ALSFRS-r, progression rate [13], and predominant upper or lower motor neuron were used as covariates. Finally, 11 ALS-RIL patients were retained for the comparison with 11 ALS-EDA patients at T6.
(b) The second step of the study included a longitudinal analysis comparing paired MRI findings in ALS-EDA at T6 vs T0.
Cross-sectional MRI analysis
The group differences of CT between ALS-EDA at T0 and CS and between ALS-EDA at T6 and ALS-RIL were explored with two types of analysis: a vertex-based analysis and a region of interest (ROI)-based one. The whole-brain vertex-wise analysis is a point-by-point group comparison of thickness across the cortical surface, without any a priori hypothesis, starting with the average images of each group. This statistical analysis was performed using Qdec (Query, Design, Estimate and Contrast), a module of Freesurfer, developed to design and execute surface analysis using age and gender as covariates. To correct for multiple comparisons, we performed Monte-Carlo cluster-based simulation with 10.000 permutations and we searched for significant clusters with p value level 0.05[22]. Areas showing significant cortical thinning were superimposed on the template. The vertex-analyses were supplemented by a ROI analysis, using the mean values of CT in the primary motor area (bilateral precentral gyrus).
WM analysis was employed on FA parameters using a two-sample t-test, with cluster-based correction for multiple comparisons (p value = 0.05) using Threshold Free Cluster-Enhancement (TFCE) correction [23]. Johns Hopkins white matter DTI-based atlas (available in the FSL software) was employed to identify WM tracts with abnormal findings and to extract their FA mean values. Between groups comparison was assessed using the non-parametric Mann–Whitney U test.
Longitudinal MRI analysis
The longitudinal analysis compared T6 vs T0 paired MRI findings in ALS-EDA patients. Because all subjects have the same number of time points, symmetrized percent change (SPC), which is a dimensionless measure of change, was computed at each vertex for each participant to assess percent change [18]. SPC is defined according to the subsequent formula: SPC = 100 × (V2 − V1) 0.5 × (V1 + V2), where V1 is the vertex-wise brain measure at T0 and V2 is the measure at 6-month follow-up. SPC maps of CT were computed for each subject. Finally, one-sample group mean-test was performed to test if the symmetrized percent change in our sample is different from zero. Results were corrected for multiple comparisons with a two-tailed permutation simulation [24], using a cluster-wise forming threshold of p < 0.05 and 10 000 random permutations. Maps were visualized by overlaying significant clusters on top of the cortical surface in the visualization tool Freeview. Longitudinal TBSS protocol was performed using a paired two-sample t-test with TFCE [23] correction (alpha = 0.05) and the results were projected to FA MNI 152 template. We then applied a mask on the results to extract bilateral corticospinal tract (CST) mean FA. Within group comparisons were performed using a Wilcoxon signed-rank test.
Sample size calculation
A minimum sample size was calculated to assess the effect of edaravone on CT of precentral gyrus and FA of CST. A free software available at http://hedwig.mgh.harvard.edu/sample_size/size.html was used for sample size calculation. To detect a mean difference of 0.25 mm in CT of precentral gyrus, as previously indicated in literature [25], with a standard deviation of 0.13 [26], a total of 12 patients had to be entered in this two treatment parallel-design study, considering a power of 80%, a type I error rate of 0.05 and two-sided analyses. Regarding the effect of edaravone on FA of CST, to our knowledge, there are not cross-sectional studies evaluating the minimal detectable change of FA of CST in two compared groups of ALS patients. Considering that the propensity score-matching method limited our analysis to 22 MRI examination, the minimal difference in FA of CST detectable by our study was 0.038, with a 0.03 standard deviation of CST [27], a power of 80%, a type I error rate of 0.05 and two-sided analyses.
Results
Clinical and demographic features of ALS-EDA at T0, CS, ALS-EDA at T6 and matched ALS-RIL are summarized in Table 1
Cross-sectional MRI analysis
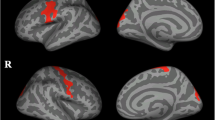
ALS-EDA at T0 showed a widespread cortical thinning in the left hemisphere, in particular in the precentral, pericalcarine and inferior-parietal cortex, and in the right hemisphere, in the precentral and paracentral cortex compared to MRI findings of CS (Fig. 1). The ROI analysis revealed significant cortical thinning in right (p = 0.019) and left (p = 0.016) precentral gyrus. Similarly, TBSS analysis revealed in ALS-EDA a reduction FA of bilateral CST from the cortex to the cerebral peduncles, more relevant in the left hemisphere (Fig. 2). Probabilistic tractography analysis confirmed the significant reduction of FA in bilateral CST (right CST: p = 0.035, left CST p = 0.002) in comparison to CS (Table 2).
Vertex-wise analysis of CT in ALS-EDA patients related to CS. ALS-EDA revealed cortical thinning in the right precentral and paracentral cortex (a) and in the left precentral, pericalcarine and inferior-parietal cortex (b). The coloured areas represented the significance maps after Monte Carlo cluster-wise correction
The comparison between ALS-EDA at T6 and ALS-RIL did not show differences in cortical thickness and FA of the CST bilaterally (Table 2).
Longitudinal MRI analysis
When compared to ALS-EDA at T6 vs T0, SPC of CT revealed one significant cluster in the bilateral precentral cortex with predominant impairment in the right hemisphere (cluster size: left: 1659.46 mm2, right: 2774.77 mm2) (Fig. 3). WM analysis showed a significant widespread reduction of FA in several white matter tracts, in particular, in anterior commissure, arcuate fasciculus, rostrum of corpus callosum and bilateral corticospinal pathways (Fig. 4). After ROI analysis, a significant reduction of FA values was found bilaterally in the CST (left hemisphere, p = 0.033; and right hemisphere, p = 0.041).
Symmetrized percent change maps of ALS-EDA patients after 6 months of treatment. Longitudinal analysis revealed cortical thinning in the bilateral precentral cortex in ALS-EDA after 6 months of treatment with edaravone in add on to riluzole. Coloured areas represented the significance maps after cluster-wise correction with a two-tailed permutation simulation in right hemisphere (a), and in left hemisphere (b)
Longitudinal TBSS analysis of ALS-EDA patients after 6-months of follow-up. Areas with reduced FA are shown in yellow–red in anterior commissure, arcuate fasciculus, rostrum of corpus callosum and bilateral CST of ALS-EDA at T6. Regions of significant differences are overlaid on axial (a), coronal (b) and sagittal (c) of the MNI152 1 mm template
Discussion
MRI generated measures have been proposed as potential diagnostic, prognostic biomarkers of ALS, nevertheless, to date, there are no published studies on MRI measures as an indicator of the effectiveness of drugs used in the disease. In this pioneering study, we have tried to investigate baseline and six-month MRI changes in real life on newly diagnosed ALS patients treated with edaravone, using CT and DTI analysis.
At baseline, the comparison between ALS patients and control subjects showed the typical pattern of ALS pathology in CT of both primary motor areas and in FA of both corticospinal. Almost all the studies carried out with CT analysis showed a predominant thinning of the precentral gyrus of ALS patients, confirming that the primary motor cortex is a distinctive site of microstructural alterations that characterizes the disease [28]. Likewise, DTI studies found alterations of the corticospinal tracts, which appears rapidly, already in the early stages of the disease, due to damage to the neurolemma or the myelin sheath [29]. The homogeneity of the results of the cross-sectional studies suggested the use of MRI as an optimal biomarker of ALS pathology [30].
Our objective was to evaluate the effect of edaravone on MRI measures and, for this purpose, MRI examination has been repeated after six months which, although short, a proportionate time considering the rapid course of the disease. Comparing baseline with follow-up MRI metrics, the most relevant results are that, at six months, CT analysis showed further cortical thinning in the motor areas. Similarly, TBSS showed a greater involvement of corticospinal tracts, but also a widespread involvement of other strategical traits of the ALS pathology [31]. Differently from MRI cross-sectional literature data, longitudinal studies did not report univocal results [32,33,34]. It seems that the discrepancy of the data reflect the heterogeneity of the progression of the disease [10]. To evaluate structural changes during the course of the disease, it is necessary to select clinically homogeneous groups. Our study group, although small, has the advantage of being representative of an early stage of disease, due to the restrictive criteria of the enrolment.
The widespread involvement of white matter tracts that we found at T6 includes the anterior commissure, the rostrum of the corpus callosum and the nearest frontal tracts. This pattern could be a confirmation of the prion-like hypothesis of disease progression, through the bi-hemispheric connections [35]. In this regard, an important emphasis has been given to the role of the corpus callosum in ALS. Histopathological evidences and more recent DTI and voxel-based morphometry studies have all confirmed a degeneration of the corpus callosum in ALS [36,37,38,39].
In the planning phase of the study, we ran into the problem of defining the ALS group to compare with ALS-EDA, because, for ethical reasons, we could not avoid administering edaravone to patients who could benefit from it. To overcome this obstacle, we used PSM to extract from our ALS database a group of matched patients taking only riluzole to compare with ALS-EDA group. To date, several studies used this method to obtain a matched sample with similar MRI measures [40,41,42]. Our results did not show evidence of a difference between edaravone and riluzole. This evidence is in agreement with clinical data by the Italian EDARAVALS Study Group, which reported no benefit of this drug in terms of disease progression and respiratory function [9].
One limit of our single centre real life study is the small sample size. However, as reported in sample size calculation section, the number of recruited patients is widely sufficient to detect difference in CT of precentral gyrus as previously reported in literature. To our knowledge, no data of a minimal detectable change of the FA of CST are available in two compared cross-sectionally ALS groups of patients. One study conducted in stroke patients reported 0.015 as the minimal detectable change in FA of CST, using the probabilistic tract approach [43]. The minimal difference found in our study is still adequate in our opinion, considering that this value was obtained in a non-easily reproducible setting with a widespread disease.
Another possible limit is the short time of follow-up; however, we feel that these did not bias our results, indicating that patients treated with edaravone showed progression of damage in the motor cortex and several WM tracts.
In our opinion, this study might provide a methodical approach that could be applied in other pharmacological clinical studies in ALS disease.
Data availability
The data that support the findings of this study are available from the corresponding author, [ILS], upon reasonable request.
Code availability
Not applicable.
References
Chiò A, Moglia C, Canosa A et al (2019) Cognitive impairment across ALS clinical stages in a population-based cohort. Neurology 93(10):e984–e994. https://doi.org/10.1212/WNL.0000000000008063
D’Amico E, Factor-Litvak P, Santella RM, Mitsumoto H (2013) Clinical perspective on oxidative stress in sporadic amyotrophic lateral sclerosis. Free Radic Biol Med 65:509–527. https://doi.org/10.1016/j.freeradbiomed.2013.06.029
Writing Group, Edaravone (MCI-186) ALS 19 Study Group (2017) Safety and efficacy of edaravone in well-defined patients with amyotrophic lateral sclerosis: a randomised, double-blind, placebo-controlled trial. Lancet Neurol 16(7):505–512. https://doi.org/10.1016/S1474-4422(17)30115-1
Akimoto M, Nakamura K, Writing Group on behalf of the Edaravone (MCI-186) ALS 19 Study Group (2017) Edaravone for treatment of early-stage ALS - Authors’ reply. Lancet Neurol 16(10):772. https://doi.org/10.1016/S1474-4422(17)30290-9
Shefner J, Heiman-Patterson T, Pioro EP et al (2020) Long-term edaravone efficacy in amyotrophic lateral sclerosis: Post-hoc analyses of Study 19 (MCI186-19). Muscle Nerve 61(2):218–221. https://doi.org/10.1002/mus.26740
Fortuna A, Gizzi M, Bello L et al (2019) Safety and efficacy of edaravone compared to historical controls in patients with amyotrophic lateral sclerosis from North-Eastern Italy. J Neurol Sci 404:47–51. https://doi.org/10.1016/j.jns.2019.06.006
Abraham A, Nefussy B, Fainmesser Y, Ebrahimi Y, Karni A, Drory VE (2019) Early post-marketing experience with edaravone in an unselected group of patients with ALS. Amyotroph Lateral Scler Frontotemporal Degener 20(3–4):260–263. https://doi.org/10.1080/21678421.2019.1572191
Okada M, Yamashita S, Ueyama H, Ishizaki M, Maeda Y, Ando Y (2018) Long-term effects of edaravone on survival of patients with amyotrophic lateral sclerosis. eNeurological Sci 11:11–14. https://doi.org/10.1016/j.ensci.2018.05.001
Lunetta C, Moglia C, Lizio A et al (2020) The Italian multicenter experience with edaravone in amyotrophic lateral sclerosis. J Neurol 267(11):3258–3267. https://doi.org/10.1007/s00415-020-09993-z
Menke RAL, Agosta F, Grosskreutz J, Filippi M, Turner MR (2017) Neuroimaging endpoints in amyotrophic lateral sclerosis. Neurotherapeutics 14(1):11–23. https://doi.org/10.1007/s13311-016-0484-9
Ferraro PM, Agosta F, Riva N et al (2017) Multimodal structural MRI in the diagnosis of motor neuron diseases. Neuroimage Clin 16:240–247. https://doi.org/10.1016/j.nicl.2017.08.002
Brooks BR (1994) El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis. Subcommittee on Motor Neuron Diseases/Amyotrophic Lateral Sclerosis of the World Federation of Neurology Research Group on Neuromuscular Diseases and the El Escorial “Clinical limits of amyotrophic lateral sclerosis” workshop contributors. J Neurol Sci 124:96–107. https://doi.org/10.1016/0022-510x(94)90191-0
Kimura F, Fujimura C, Ishida S et al (2006) Progression rate of ALSFRS-R at time of diagnosis predicts survival time in ALS. Neurology 66(2):265–267. https://doi.org/10.1212/01.wnl.0000194316.91908.8a
Quinn C, Edmundson C, Dahodwala N, Elman L (2020) Reliable and efficient scale to assess upper motor neuron disease burden in amyotrophic lateral sclerosis. Muscle Nerve 61(4):508–511. https://doi.org/10.1002/mus.26764
Grachev ID, Berdichevsky D, Rauch SL et al (1999) A method for assessing the accuracy of intersubject registration of the human brain using anatomic landmarks. Neuroimage 9(2):250–268. https://doi.org/10.1006/nimg.1998.0397
Reuter M, Rosas HD, Fischl B (2010) Highly accurate inverse consistent registration: a robust approach. Neuroimage 53(4):1181–1196. https://doi.org/10.1016/j.neuroimage.2010.07.020
Reuter M, Fischl B (2011) Avoiding asymmetry-induced bias in longitudinal image processing. Neuroimage 57(1):19–21. https://doi.org/10.1016/j.neuroimage.2011.02.076
Reuter M, Schmansky NJ, Rosas HD, Fischl B (2012) Within-subject template estimation for unbiased longitudinal image analysis. Neuroimage 61(4):1402–1418. https://doi.org/10.1016/j.neuroimage.2012.02.084
Smith SM, Jenkinson M, Johansen-Berg H et al (2006) Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage 31(4):1487–1505. https://doi.org/10.1016/j.neuroimage.2006.02.024
Menke RAL, Körner S, Filippini N et al (2014) Widespread grey matter pathology dominates the longitudinal cerebral MRI and clinical landscape of amyotrophic lateral sclerosis. Brain 137(Pt 9):2546–2555. https://doi.org/10.1093/brain/awu162
Austin PC (2007) Propensity-score matching in the cardiovascular surgery literature from 2004 to 2006: a systematic review and suggestions for improvement. J Thorac Cardiovasc Surg 134(5):1128–1135. https://doi.org/10.1016/j.jtcvs.2007.07.021
Hutton C, Draganski B, Ashburner J, Weiskopf N (2009) A comparison between voxel-based cortical thickness and voxel-based morphometry in normal aging. Neuroimage 48(2):371–380. https://doi.org/10.1016/j.neuroimage.2009.06.043
Smith SM, Nichols TE (2009) Threshold-free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage 44(1):83–98. https://doi.org/10.1016/j.neuroimage.2008.03.061
Greve DN, Fischl B (2018) False positive rates in surface-based anatomical analysis. Neuroimage 171:6–14. https://doi.org/10.1016/j.neuroimage12.072
Pardoe HR, Abbott DF, Jackson GD, Initiative ADN (2013) Sample size estimates for well-powered cross-sectional cortical thickness studies. Hum Brain Mapp 34(11):3000–3009. https://doi.org/10.1002/hbm.22120
Mezzapesa DM, D’Errico E, Tortelli R et al (2013) Cortical thinning and clinical heterogeneity in amyotrophic lateral sclerosis. PLoS ONE 8(11):e80748. https://doi.org/10.1371/journal.pone.0080748
Spinelli EG, Riva N, Rancoita PMV et al (2020) Structural MRI outcomes and predictors of disease progression in amyotrophic lateral sclerosis. Neuroimage Clin 27:102315. https://doi.org/10.1016/j.nicl.2020.102315
Grolez G, Moreau C, Danel-Brunaud V et al (2016) The value of magnetic resonance imaging as a biomarker for amyotrophic lateral sclerosis: a systematic review. BMC Neurol 16(1):155. https://doi.org/10.1186/s12883-016-0672-6
Ellis CM, Simmons A, Jones DK, Bland J, Dawson JM, Horsfield MA, Williams SC, Leigh PN (1999) Diffusion tensor MRI assesses corticospinal tract damage in ALS. Neurology 53(5):1051–1058. https://doi.org/10.1212/wnl.53.5.1051 (PMID: 10496265)
Mazón M, Vázquez Costa JF, Ten-Esteve A, Martí-Bonmatí L (2018) Imaging biomarkers for the diagnosis and prognosis of neurodegenerative diseases. The example of amyotrophic lateral sclerosis. Front Neurosci. https://doi.org/10.3389/fnins.2018.00784
Van der Graaff MM, Sage CA, Caan MWA et al (2011) Upper and extra-motoneuron involvement in early motoneuron disease: a diffusion tensor imaging study. Brain 134(Pt 4):1211–1228. https://doi.org/10.1093/brain/awr016
Agosta F, Rocca MA, Valsasina P et al (2009) A longitudinal diffusion tensor MRI study of the cervical cord and brain in amyotrophic lateral sclerosis patients. J Neurol Neurosurg Psychiatry 80(1):53–55. https://doi.org/10.1136/jnnp.2008.154252
Keil C, Prell T, Peschel T, Hartung V, Dengler R, Grosskreutz J (2012) Longitudinal diffusion tensor imaging in amyotrophic lateral sclerosis. BMC Neurosci 13:141. https://doi.org/10.1186/1471-2202-13-141
Schuster C, Kasper E, Machts J et al (2014) Longitudinal course of cortical thickness decline in amyotrophic lateral sclerosis. J Neurol 261(10):1871–1880. https://doi.org/10.1007/s00415-014-7426-4
Eisen A, Braak H, Del Tredici K, Lemon R, Ludolph AC, Kiernan MC (2017) Cortical influences drive amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry 88(11):917–924. https://doi.org/10.1136/jnnp-2017-315573
Smith MC (1960) Nerve fibre degeneration in the brain in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry 23(4):269–282. https://doi.org/10.1136/jnnp.23.4.269
Yamauchi H, Fukuyama H, Ouchi Y et al (1995) Corpus callosum atrophy in amyotrophic lateral sclerosis. J Neurol Sci 134(1–2):189–196. https://doi.org/10.1016/0022-510x(95)00220-6
Filippini N, Douaud G, Mackay CE, Knight S, Talbot K, Turner MR (2010) Corpus callosum involvement is a consistent feature of amyotrophic lateral sclerosis. Neurology 75(18):1645–1652. https://doi.org/10.1212/WNL.0b013e3181fb84d1
Kim J-E, Oh JS, Sung J-J, Lee K-W, Song IC, Hong Y-H (2014) Diffusion tensor tractography analysis of the corpus callosum fibers in amyotrophic lateral sclerosis. J Clin Neurol 10(3):249–256. https://doi.org/10.3988/jcn.2014.10.3.249
Chung SJ, Choi Y-H, Kwon H et al (2017) Sleep disturbance may alter white matter and resting state functional connectivities in Parkinson’s disease. Sleep. https://doi.org/10.1093/sleep/zsx009
Ritchie SJ, Dickie DA, Cox SR et al (2018) Brain structural differences between 73- and 92-year olds matched for childhood intelligence, social background, and intracranial volume. Neurobiol Aging 62:146–158. https://doi.org/10.1016/j.neurobiolaging.2017.10.005
Kline RP, Pirraglia E, Cheng H et al (2012) Surgery and Brain Atrophy In Cognitively Normal Elderly Subjects and Subjects Diagnosed with Mild Cognitive Impairment. Anesthesiology 116(3):603–612. https://doi.org/10.1097/ALN.0b013e318246ec0b
Lewis AF, Myers M, Heiser J, Kolar M, Baird JF (2020) Stewart JC (2020) Test-retest reliability and minimal detectable change of corticospinal tract integrity in chronic stroke. Hum Brain Mapp 41(9):2514–2526. https://doi.org/10.1002/hbm.24961
Funding
Open access funding provided by Università degli Studi di Bari Aldo Moro within the CRUI-CARE Agreement.. The authors did not receive support from any organization for the submitted work. No funding was received to assist with the preparation of this manuscript. No funding was received for conducting this study. No funds, grants, or other support was received.
Author information
Authors and Affiliations
Contributions
ILS and DM conceptualized the study, had full access to all data and take responsibility for the integrity of the data and the accuracy of the data analysis. ED and GM interpreted clinical and MRI data. ED, GM and AI contributed to the data analysis and the writing of the manuscript. All the authors contributed to the data interpretation and reviewed and approved the final version. Authors declare that the work described has not been published before; that it is not under consideration for publication anywhere else; that its publication has been approved by all co-authors, if any, as well as by the responsible authorities—tacitly or explicitly—at the institute where the work has been carried out.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines. The study was approved by the Local Independent Ethics Committee of the Regional University Hospital Polyclinic of Bari, Italy.
Consent to participate
All patients gave written informed consent to be enrolled in the study.
Consent for publication
All patients gave written informed consent to publish data for scientific purpose.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Distaso, E., Milella, G., Mezzapesa, D.M. et al. Magnetic resonance metrics to evaluate the effect of therapy in amyotrophic lateral sclerosis: the experience with edaravone. J Neurol 268, 3307–3315 (2021). https://doi.org/10.1007/s00415-021-10495-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00415-021-10495-9