Abstract
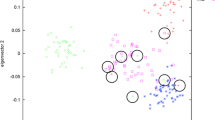
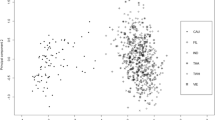
Ancestry informative markers (AIMs) can be used to detect and adjust for population stratification and predict the ancestry of the source of an evidence sample. Autosomal single nucleotide polymorphisms (SNPs) are the best candidates for AIMs. It is essential to identify the most informative AIM SNPs across relevant populations. Several informativeness measures for ancestry estimation have been used for AIMs selection: absolute allele frequency differences (δ), F statistics (F ST), and informativeness for assignment measure (In). However, their efficacy has not been compared objectively, particularly for determining affiliations of major US populations. In this study, these three measures were directly compared for AIMs selection among four major US populations, i.e., African American, Caucasian, East Asian, and Hispanic American. The results showed that the F ST panel performed slightly better for population resolution based on principal component analysis (PCA) clustering than did the δ panel and both performed better than the In panel. Therefore, the 23 AIMs selected by the F ST measure were used to characterize the four major American populations. Genotype data of nine sample populations were used to evaluate the efficiency of the 23-AIMs panel. The results indicated that individuals could be correctly assigned to the major population categories. Our AIMs panel could contribute to the candidate pool of AIMs for potential forensic identification purposes.








Similar content being viewed by others
References
Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW (2002) Genetic structure of human populations. Science 298:2381–2385
Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM (2003) Control of confounding of genetic associations in stratified populations. Am J Hum Genet 72:1492–1504
Shriver MD, Parra EJ, Dios S, Bonilla C, Norton H, Jovel C, Pfaff C, Jones C, Massac A, Cameron N, Baron A, Jackson T, Argyropoulos G, Jin L, Hoggart CJ, McKeigue PM, Kittles RA (2003) Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet 112:387–399
Marchini J, Cardon LR, Phillips MS, Donnelly P (2004) The effects of human population structure on large genetic association studies. Nat Genet 36:512–517
Jobling MA, Gill P (2004) Encoded evidence: DNA in forensic analysis. Nat Rev Genet 5:739–751
Yang N, Li H, Criswell LA, Gregersen PK, Alarcon-Riquelme ME, Kittles R, Shigeta R, Silva G, Patel PI, Belmont JW, Seldin MF (2005) Examination of ancestry and ethnic affiliation using highly informative diallelic DNA markers: application to diverse and admixed populations and implications for clinical epidemiology and forensic medicine. Hum Genet 118:382–392
Shriver MD, Kittles RA (2004) Genetic ancestry and the search for personalized genetic histories. Nat Rev Genet 5:611–618
King JL, LaRue BL, Novroski NM, Stoljarova M, Seo SB, Zeng X, Warshauer DH, Davis CP, Parson W, Sajantila A, Budowle B (2014) High-quality and high throughput massively parallel sequencing of the human mitochondrial genome using the Illumina MiSeq. Forensic Sci Int Genet 12:128–135
Jobling MA, Tyler-Smith C (2003) The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 4:598–612
Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC (1991) African populations and the evolution of human mitochondrial DNA. Science 253:1503–1507
Hammond HA, Jin L, Zhong Y, Caskey CT, Chakraborty R (1994) Evaluation of 13 short tandem repeat loci for use in personal identification applications. Am J Hum Genet 55:175–189
Jin L, Chakraborty R (1995) Population structure, stepwise mutations, heterozygote deficiency and their implications in DNA forensics. Heredity 74:274–285
Smith MW, Lautenberger JA, Shin HD, Chretien JP, Shrestha S, Gilbert DA, O’Brien SJ (2001) Markers for mapping by admixture linkage disequilibrium in African American and Hispanic populations. Am J Hum Genet 69:1080–1094
Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29:308–311
International HapMap Consortium (2003) The International HapMap Project. Nature 426:789–796
1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65
Phillips C, Salas A, Sánchez JJ, Fondevila M, Gómez-Tato A, Alvarez-Dios J, Calaza M, de Cal MC, Ballard D, Lareu MV, Carracedo A, SNPforID Consortium (2007) Inferring ancestral origin using a single multiplex assay of ancestry-informative marker SNPs. Forensic Sci Int Genet 1:273–280
Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, Kittles R, Alarcon-Riquelme ME, Gregersen PK, Belmont JW, De La Vega FM, Seldin MF (2009) Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat 30:69–78
Kidd KK, Speed WC, Pakstis AJ, Furtado MR, Fang R, Madbouly A, Maiers M, Middha M, Friedlaender FR, Kidd JR (2014) Progress toward an efficient panel of SNPs for ancestry inference. Forensic Sci Int Genet 10:23–32
Nievergelt CM, Maihofer AX, Shekhtman T, Libiger O, Wang X, Kidd KK, Kidd JR (2013) Inference of human continental origin and admixture proportions using a highly discriminative ancestry informative 41-SNP panel. Investig Genet 4:13
Wei YL, Wei L, Zhao L, Sun QF, Jiang L, Zhang T, Liu HB, Chen JG, Ye J, Hu L, Li CX (2015) A single-tube 27-plex SNP assay for estimating individual ancestry and admixture from three continents. Int J Legal Med
Rosenberg NA, Li LM, Ward R, Pritchard JK (2003) Informativeness of genetic markers for inference of ancestry. Am J Hum Genet 73:1402–1422
Wright S (1950) Genetical structure of populations. Nature 166:247–249
Ding L, Wiener H, Abebe T, Altaye M, Go RC, Kercsmar C, Grabowski G, Martin LJ, Khurana Hershey GK, Chakorborty R, Baye TM (2011) Comparison of measures of marker informativeness for ancestry and admixture mapping. BMC Genomics 12:622
Amirisetty S, Hershey GK, Baye TM (2012) AncestrySNPminer: a bioinformatics tool to retrieve and develop ancestry informative SNP panels. Genomics 100:57–63
Lewis PO, Zaykin D (2001) Genetic Data Analysis: computer program for the analysis of allelic data. Version 1.0 (d16c). http://hydrodictyon.eeb.uconn.edu/people/plewis/software.php. Accessed 25 April 2007.
Patterson N, Price AL, Reich D (2006) Population structure and eigenanalysis. PLoS Genet 2:e190
Zweig MH, Campbell G (1993) Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem 39:561–577
Qin P, Li Z, Jin W, Lu D, Lou H, Shen J, Jin L, Shi Y, Xu S (2014) A panel of ancestry informative markers to estimate and correct potential effects of population stratification in Han Chinese. Eur J Hum Genet 22:248–253
Adinsoft SARL (2010) XLSTAT-software. Version 10. Addinsoft, Paris
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
SPSS Inc (2007) SPSS for Windows. Version 16.0. Chicago
Green SB, Salkind NJ, Akey TM (2008) Using SPSS for Windows and Macintosh: analyzing and understanding data. Prentice Hall, New Jersey
Kidd JM, Gravel S, Byrnes J, Moreno-Estrada A, Musharoff S, Bryc K, Degenhardt JD, Brisbin A, Sheth V, Chen R, McLaughlin SF, Peckham HE, Omberg L, Bormann-Chung CA, Stanley S, Pearlstein K, Levandowsky E, Gravel S, Acevedo-Acevedo S, Auton A, Keinan A, Acuna-Alonzo V, Canizales-Quinteros S, Eng C, Burchard EG, Russell A, Reynolds A, Clark AG, Reese M, Lincoln SE, Butte AJ, De La Vega FM, Bustamante CD (2012) Population genetic inference from personal genome data: impact of ancestry and admixture on human genomic variation. Am J Hum Genet 91:660–671
Wall JD, Jiang R, Gignoux C, Chen GK, Eng C, Huntsman S, Marjoram P (2011) Genetic variation in Native Americans, inferred from Latino SNP and resequencing data. Mol Biol Evol 28:2231–2237
Salazar-Flores J, Zuñiga-Chiquette F, Rubi-Castellanos R, Álvarez-Miranda JL, Zetina-Hérnandez A, Martínez-Sevilla VM, González-Andrade F, Corach D, Vullo C, Álvarez JC, Lorente JA, Sánchez-Diz P, Herrera RJ, Cerda-Flores RM, Muñoz-Valle JF, Rangel-Villalobos H (2015) Admixture and genetic relationships of Mexican Mestizos regarding Latin American and Caribbean populations based on 13 CODIS-STRs. Homo 66:44–59
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Rosenberg N (2004) Distruct: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Bushnell D, Hudson RA (2010) Colombia: a country study. Federal Research Division, Library of Congress, Washingtion D.C
Halder I, Shriver M, Thomas M, Fernandez JR, Frudakis T (2008) A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: utility and applications. Hum Mutat 29:648–658
Phillips C, Parson W, Lundsberg B, Santos C, Freire-Aradas A, Torres M, Eduardoff M, Børsting C, Johansen P, Fondevila M, Morling N, Schneider P, EUROFORGEN-NoE Consortium, Carracedo A, Lareu MV (2014) Building a forensic ancestry panel from the ground up: the EUROFORGEN Global AIM-SNP set. Forensic Sci Int Genet 11:13–25
Gettings KB, Lai R, Johnson JL, Peck MA, Hart JA, Gordish-Dressman H, Schanfield MS, Podini DS (2014) A 50-SNP assay for biogeographic ancestry and phenotype prediction in the US population. Forensic Sci Int Genet 8:101–108
Jia J, Wei YL, Qin CJ, Hu L, Wan LH, Li CX (2014) Developing a novel panel of genome-wide ancestry informative markers for bio-geographical ancestry estimates. Forensic Sci Int Genet 8:187–194
Rogalla U, Rychlicka E, Derenko MV, Malyarchuk BA, Grzybowski T (2015) Simple and cost-effective 14-loci SNP assay designed for differentiation of European, East Asian and African samples. Forensic Sci Int Genet 14:42–49
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Table 1
The top AIMs selected by three measures from ASW and CEU after H-W and LD selection. The top thirty SNPs were reduced to 26, 26, 26 AIMs by δ, F ST, and In, respectively. 1–3 indicated the SNPs that were in LD, and the highlighted ones (Bold and Italic) were removed (XLSX 12 kb)
Supplemental Table 2
The top AIMs selected by three measures from ASW and CHD after H-W and LD selection. The top thirty SNPs were reduced to 24, 25, 26 AIMs by δ, F ST, and In, respectively. 1–5 indicated the SNPs that were in LD, and the highlighted ones (Bold and Italic) were removed (XLSX 13 kb)
Supplemental Table 3
The top AIMs selected by three measures from ASW and MEX after H-W and LD selection. The top thirty SNPs were reduced to 27, 26, 25 AIMs by δ, F ST, and In, respectively. 1–4 indicated the SNPs that were in LD, and the highlighted ones (Bold and Italic) were removed (XLSX 13 kb)
Supplemental Table 4
The top AIMs selected by three measures from CEU and CHD after H-W and LD selection. The top thirty SNPs were reduced to 27, 27, 29 AIMs by δ, F ST, and In, respectively. 1–3 indicated the SNPs that were in LD, and the highlighted ones (Bold and Italic) were removed (XLSX 14 kb)
Supplemental Table 5
The top AIMs selected by three measures from CEU and MEX after H-W and LD selection. The top thirty SNPs were reduced to 26, 24, 22 AIMs by δ, F ST, and In, respectively. 1–6 indicated the SNPs that were in LD, and the highlighted ones (Bold and Italic) were removed (XLSX 13 kb)
Supplemental Table 6
The top AIMs selected by three measures from CHD and MEX after H-W and LD selection. The top thirty SNPs were reduced to 22, 24, 24 AIMs by δ, F ST, and In, respectively. 1–5 indicated the SNPs that were in LD, and the highlighted ones (Bold and Italic) were removed (XLSX 13 kb)
Supplemental Table 7
The minimum number of markers to distinguish any two populations identified by three measures (δ, F ST and In) (XLSX 12 kb)
Supplemental Table 8
The correlation coefficients of PC1 and PC2 values among δ, F ST and In panels (XLSX 10 kb)
Supplemental Table 9
Ancestry prediction of HapMap individuals that fell outside the 95 % confidence interval of four major US populations (XLSX 13 kb)
Supplemental Table 10
Ancestry prediction of 1000 Genomes individuals that fell outside the 95 % confidence interval of four major US populations. All CLM individuals were listed, because CLM contains individuals from three populations (XLSX 20 kb)
Supplemental Table 11
Summary of SNPs contained in ten AIMs panels (XLSX 29 kb)
Rights and permissions
About this article
Cite this article
Zeng, X., Chakraborty, R., King, J.L. et al. Selection of highly informative SNP markers for population affiliation of major US populations. Int J Legal Med 130, 341–352 (2016). https://doi.org/10.1007/s00414-015-1297-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-015-1297-9