Abstract
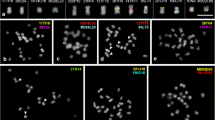
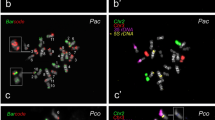
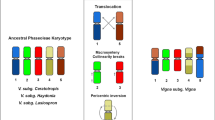
Cytogenomic resources have accelerated synteny and chromosome evolution studies in plant species, including legumes. Here, we established the first cytogenetic map of V. angularis (Va, subgenus Ceratotropis) and compared this new map with those of V. unguiculata (Vu, subgenus Vigna) and P. vulgaris (Pv) by BAC-FISH and oligopainting approaches. We mapped 19 Vu BACs and 35S rDNA probes to the 11 chromosome pairs of Va, Vu, and Pv. Vigna angularis shared a high degree of macrosynteny with Vu and Pv, with five conserved syntenic chromosomes. Additionally, we developed two oligo probes (Pv2 and Pv3) used to paint Vigna orthologous chromosomes. We confirmed two reciprocal translocations (chromosomes 2 and 3 and 1 and 8) that have occurred after the Vigna and Phaseolus divergence (~9.7 Mya). Besides, two inversions (2 and 4) and one translocation (1 and 5) have occurred after Vigna and Ceratotropis subgenera separation (~3.6 Mya). We also observed distinct oligopainting patterns for chromosomes 2 and 3 of Vigna species. Both Vigna species shared similar major rearrangements compared to Pv: one translocation (2 and 3) and one inversion (chromosome 3). The sequence synteny identified additional inversions and/or intrachromosomal translocations involving pericentromeric regions of both orthologous chromosomes. We propose chromosomes 2 and 3 as hotspots for chromosomal rearrangements and de novo centromere formation within and between Vigna and Phaseolus. Our BAC- and oligo-FISH mapping contributed to physically trace the chromosome evolution of Vigna and Phaseolus and its application in further studies of both genera.




Similar content being viewed by others
Data availability
All data generated during this study are available in this published paper and its Supplementary Information files.
References
Albert PS, Zhang T, Semrau K, Rouillard JM, Kao YH, Wang CJR, Danilova TV, Jiang J, Birchler JA (2019) Whole-chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships. Proc Natl Acad Sci 116:1679–1685. https://doi.org/10.1073/pnas.1813957116
Almeida C, Pedrosa-Harand A (2013) High macro-collinearity between lima bean (Phaseolus lunatus L.) and the common bean (P. vulgaris L.) as revealed by comparative cytogenetic mapping. Theor Appl Genet 126:1909–1916. https://doi.org/10.1007/s00122-013-2106-9
Beliveau BJ, Joyce EF, Apostolopoulos N, Yilmaz F, Fonseka CY, McCole RB, Chang Y, Li JB, Senaratne TN, Williams BR, Rouillard JM, Wu CT (2012) Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc Natl Acad Sci 109:21301–21306. https://doi.org/10.1073/pnas.1213818110
Betekhtin A, Jenkins G, Hasterok R (2014) Reconstructing the evolution of Brachypodium genomes using comparative chromosome painting. PLoS One 9:e115108. https://doi.org/10.1371/journal.pone.0115108
Bi Y, Zhao Q, Yan W et al (2019) Flexible chromosome painting based on multiplex PCR of oligonucleotides and its application for comparative chromosome analyses in Cucumis. Plant J. https://doi.org/10.1111/tpj.14600
Bonifácio EM, Fonsêca A, Almeida C, dos Santos KG, Pedrosa-Harand A (2012) Comparative cytogenetic mapping between the lima bean (Phaseolus lunatus L.) and the common bean (P. vulgaris L.). Theor Appl Genet 124:1513–1520. https://doi.org/10.1007/s00122-012-1806-x
Braz GT, He L, Zhao H, Zhang T, Semrau K, Rouillard JM, Torres GA, Jiang JM (2018) Comparative oligo-FISH mapping: an efficient and powerful methodology to reveal karyotypic and chromosomal evolution. Genetics 208:513–523. https://doi.org/10.1534/genetics.117.300344
Braz GT, do Vale Martins L, Zhang T, Albert PS, Birchler JA, Jiang J (2020) A universal chromosome identification system for maize and wild Zea species. Chromosom Res 28:183–194. https://doi.org/10.1007/s10577-020-09630-5
Chèvre AM, Mason AS, Coriton O, Grandont L, Jenczewski E, Lysak M (2018) Cytogenetics, a science linking genomics and breeding: the Brassica model. In: Liu S, Snowdon R, Chalhoub B (eds) The Brassica napus Genome. Compendium of Plant Genomes. Springer Nature, Switzerland, pp 21–39. https://doi.org/10.1007/978-3-319-43694-4_2
Coghlan A, Eichler EE, Oliver SG, Paterson AH, Stein L (2005) Chromosome evolution in eukaryotes: a multi-kingdom perspective. Trends Genet 21:673–682. https://doi.org/10.1016/j.tig.2005.09.009
de Carvalho CR, Saraiva LS (1993) An air drying technique for maize chromosomes without enzymatic maceration. Biotech Histochem 68:142–145. https://doi.org/10.3109/10520299309104684
Delgado-Salinas A, Thulin M, Pasquet R, Weeden N, Lavin M (2011) Vigna (Leguminosae) sensu lato: the names and identities of the American segregate genera. Am J Bot 98:1694–1715. https://doi.org/10.3732/ajb.1100069
do Vale Martins L, Yu F, Zhao H et al (2019) Meiotic crossovers characterized by haplotype-specific chromosome painting in maize. Nat Commun 10:4604. https://doi.org/10.1038/s41467-019-12646-z
Fonsêca A, Pedrosa-Harand A (2013) Karyotype stability in the genus Phaseolus evidenced by the comparative mapping of the wild species Phaseolus microcarpus. Genome 56:335–343. https://doi.org/10.1139/gen-2013-0025
Fonsêca A, Ferreira J, dos Santos TR et al (2010) Cytogenetic map of common bean (Phaseolus vulgaris L.). Chromosom Res 18:487–502. https://doi.org/10.1007/s10577-010-9129-8
Fonsêca A, Ferraz ME, Pedrosa-Harand A (2016) Speeding up chromosome evolution in Phaseolus: multiple rearrangements associated with a one-step descending dysploidy. Chromosoma 125:413–421. https://doi.org/10.1007/s00412-015-0548-3
Galasso I, Schmidt T, Pignone D, Heslop-Harrison JS (1995) The molecular cytogenetics of Vigna unguiculata (L.) Walp: the physical organization and characterization of 18s-5.8s-25s rRNA genes, 5s rRNA genes, telomere-like sequences, and a family of centromeric repetitive DNA sequences. Theor Appl Genet 91:928–935. https://doi.org/10.1007/BF00223902
Han OK, Kaga A, Isemura T, Wang XW, Tomooka N, Vaughan DA (2005) A genetic linkage map for azuki bean [Vigna angularis (Willd.) Ohwi & Ohashi]. Theor Appl Genet 111:1278–1287. https://doi.org/10.1007/s00122-005-0046-8
Han Y, Zhang Z, Liu C, Liu J, Huang S, Jiang J, Jin W (2009) Centromere repositioning in cucurbit species: implication of the genomic impact from centromere activation and inactivation. Proc Natl Acad Sci 106:14937–14941. https://doi.org/10.1073/pnas.0904833106
Han Y, Zhang T, Thammapichai P, Weng Y, Jiang J (2015) Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 200:771–779. https://doi.org/10.1534/genetics.115.177642
He L, Braz GT, Torres GA, Jiang J (2018) Chromosome painting in meiosis reveals pairing of specific chromosomes in polyploid Solanum species. Chromosoma 127:505–513. https://doi.org/10.1007/s00412-018-0682-9
He L, Zhao H, He J, Yang Z, Guan B, Chen K, Hong Q, Wang J, Liu J, Jiang J (2020) Extraordinarily conserved chromosomal synteny of Citrus species revealed by chromosome-specific painting. Plant J 103:2225–2235. https://doi.org/10.1111/tpj.14894
Heslop-Harrison JS, Harrison GE, Leitch IJ (1992) Reprobing of DNA: DNA in situ hybridization preparations. Trends Genet 8:372–373. https://doi.org/10.1016/0168-9525(92)90287-e
Hou L, Xu M, Zhang T, Xu Z, Wang W, Zhang J, Yu M, Ji W, Zhu C, Gong Z, Gu M, Jiang J, Yu H (2018) Chromosome painting and its applications in cultivated and wild rice. BMC Plant Biol 18:110. https://doi.org/10.1186/s12870-018-1325-2
Hougaard BK, Madsen LH, Sandal N, de Carvalho Moretzsohn M, Fredslund J, Schauser L, Nielsen AM, Rohde T, Sato S, Tabata S, Bertioli DJ, Stougaard J (2008) Legume anchor markers link syntenic regions between Phaseolus vulgaris, Lotus japonicus, Medicago truncatula and Arachis. Genetics 179:2299–2312. https://doi.org/10.1534/genetics.108.090084
Idziak D, Hazuka I, Poliwczak B, Wiszynska A, Wolny E, Hasterok R (2014) Insight into the karyotype evolution of Brachypodium species using comparative chromosome barcoding. PLoS One 9:e93503. https://doi.org/10.1371/journal.pone.0093503
Ishii T, Juranic M, Maheshwari S et al (2020) Unequal contribution of two paralogous CENH3 variants in cowpea centromere function. Commun Biol 3:775. https://doi.org/10.1038/s42003-020-01507-x
Iwata A, Tek AL, Richard MMS, Abernathy B, Fonsêca A, Schmutz J, Chen NWG, Thareau V, Magdelenat G, Li Y, Murata M, Pedrosa-Harand A, Geffroy V, Nagaki K, Jackson SA (2013) Identification and characterization of functional centromeres of the common bean. Plant J 76:47–60. https://doi.org/10.1111/tpj.12269
Iwata-Otsubo A, Lin JY, Gill N, Jackson SA (2016a) Highly distinct chromosomal structures in cowpea (Vigna unguiculata), as revealed by molecular cytogenetic analysis. Chromosom Res 24:197–216. https://doi.org/10.1007/s10577-015-9515-3
Iwata-Otsubo A, Radke B, Findley S, Abernathy B, Vallejos E, Jackson SA (2016b) Fluorescence in situ hybridization (FISH)-based karyotyping reveals rapid evolution of centromeric and subtelomeric repeats in common bean (Phaseolus vulgaris) and relatives. G3 (Bethesda) 6:1013–1022. https://doi.org/10.1534/g3.115.024984
Javadi F, Tun YT, Kawase M, Guan K, Yamaguchi H (2011) Molecular phylogeny of the subgenus Ceratotropis (genus Vigna, Leguminosae) reveals three eco-geographical groups and Late Pliocene-Pleistocene diversification: evidence from four plastid DNA region sequences. Ann Bot 108:367–380. https://doi.org/10.1093/aob/mcr141
Jiang J (2019) Fluorescence in situ hybridization in plants: recent developments and future applications. Chromosom Res 27:153–165. https://doi.org/10.1007/s10577-019-09607-z
Jiang J, Gill BS (2006) Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49:1057–1068. https://doi.org/10.1139/g06-076
Jiao W, Schneeberger K (2020) Chromosome-level assemblies of multiple Arabidopsis genomes reveal hotspots of rearrangements with altered evolutionary dynamics. Nat Commun 11:989. https://doi.org/10.1038/s41467-020-14779
Kang YJ, Satyawan D, Shim S, Lee T, Lee J, Hwang WJ, Kim SK, Lestari P, Laosatit K, Kim KH, Ha TJ, Chitikineni A, Kim MY, Ko JM, Gwag JG, Moon JK, Lee YH, Park BS, Varshney RK, Lee SH (2015) Draft genome sequence of adzuki bean, Vigna angularis. Sci Rep 5:8069. https://doi.org/10.1038/srep08069
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. https://doi.org/10.1101/gr.092759.109
Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL (2004) Versatile and open software for comparing large genomes. Genome Biol 5:R12. https://doi.org/10.1186/gb-2004-5-2-r12
Lewis GP (2005) Tribe Caesalpinieae. In: Lewis G, Schrire B, Mackinder B, Lock M (eds) Legumes of the world. Royal Botanic Gardens, Kew
Li H, Wang W, Lin L, Zhu X, Li J, Zhu X, Chen Z (2013) Diversification of the phaseoloid legumes: effects of climate change, range expansion and habit shift. Front Plant Sci 4:386. https://doi.org/10.3389/fpls.2013.00386
Liu X, Sun S, Wu Y, Zhou Y, Gu S, Yu H, Yi C, Gu M, Jiang J, Liu B, Zhang T, Gong Z (2019) Dual-color oligo-FISH can reveal chromosomal variations and evolution in Oryza species. Plant J 101:112–121. https://doi.org/10.1111/tpj.14522
Liu Y, Wang X, Wei Y, Liu Z, Lu Q, Liu F, Zhang T, Peng R (2020) Chromosome painting based on bulked oligonucleotides in cotton. Front Plant Sci 11:802. https://doi.org/10.3389/fpls.2020.00802
Lonardi S, Muñoz-Amatriaín M, Liang Q, Shu S, Wanamaker SI, Lo S, Tanskanen J, Schulman AH, Zhu T, Luo MC, Alhakami H, Ounit R, Hasan AM, Verdier J, Roberts PA, Santos JRP, Ndeve A, Doležel J, Vrána J, Hokin SA, Farmer AD, Cannon SB, Close TJ (2019) The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J 98:767–782. https://doi.org/10.1111/tpj.14349
LPWG (2017) A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny: the Legume Phylogeny Working Group (LPWG). Taxon 66:44–77. https://doi.org/10.12705/661.3
Lusinska J, Majka J, Betekhtin A, Susek K, Wolny E, Hasterok R (2018) Chromosome identification and reconstruction of evolutionary rearrangements in Brachypodium distachyon, B. stacei and B. hybridum. Ann Bot 122:445–459. https://doi.org/10.1093/aob/mcy086
Lysak MA, Fransz PF, Ali HB, Schubert I (2001) Chromosome painting in Arabidopsis thaliana. Plant J 28:689–697. https://doi.org/10.1046/j.1365-313x.2001.01194.x
Lysak MA, Koch MA, Pecinka A, Schubert I (2005) Chromosome triplication found across the tribe Brassiceae. Genome Res 15:516–525. https://doi.org/10.1101/gr.3531105
Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I (2006) Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci 103:5224–5229. https://doi.org/10.1073/pnas.0510791103
Mandáková T, Lysak MA (2008) Chromosomal phylogeny and karyotype evolution in x=7 crucifer species (Brassicaceae). Plant Cell 20:2559–2570. https://doi.org/10.1105/tpc.108.062166
Mandáková T, Joly S, Krzywinski M, Mummenhoff K, Lysak MA (2010) Fast diploidization in close mesopolyploid relatives of Arabidopsis. Plant Cell 22:2277–2290. https://doi.org/10.1105/tpc.110.074526
Mandáková T, Hlousková P, Koch MA, Lysak MA (2020) Genome evolution in Arabideae was marked by frequent centromere repositioning. Plant Cell 32:650–665. https://doi.org/10.1105/tpc.19.00557
Meng Z, Zhang Z, Yan T, Lin Q, Wang Y, Huang W, Huang Y, Li Z, Yu Q, Wang J, Wang K (2018) Comprehensively characterizing the cytological features of Saccharum spontaneum by the development of a complete set of chromosome-specific oligo probes. Front Plant Sci 9:1624. https://doi.org/10.3389/fpls.2018.01624
Meng Z, Han J, Lin Y, Zhao Y, Lin Q, Ma X, Wang J, Zhang M, Zhang L, Yang Q, Wang K (2020) Characterization of a Saccharum spontaneum with a basic chromosome number of x=10 provides new insights on genome evolution in genus Saccharum. Theor Appl Genet 133:187–199. https://doi.org/10.1007/s00122-019-03450-w
Mercado-Ruaro P, Delgado-Salinas A (1996) Karyological studies in several Mexican species of Phaseolus L. and Vigna Savi (Phaseolinae, Fabaceae). In: Pickersgill B, Lock JM (eds) Advances in legumes systematics. 8: Legumes of economic importance. Royal Botanic Gardens, Kew, pp 83–87
Muchero W, Diop NN, Bhat PR, Fenton RD, Wanamaker S, Pottorff M, Hearne S, Cisse N, Fatokun C, Ehlers JD, Roberts PA, Close TJ (2009) A consensus genetic map of cowpea [Vigna unguiculata (L) Walp] and synteny based on EST-derived SNPs. Proc Natl Acad Sci 106:18159–18164. https://doi.org/10.1073/pnas.0905886106
Nasuda S, Hudakova S, Schubert I, Houben A, Endo TR (2005) Stable barley chromosomes without centromeric repeats. Proc Natl Acad Sci 102:9842–9847. https://doi.org/10.1073/pnas.0504235102
Oliveira ARS, Martins LV, Bustamante FO, Muñoz-Amatriaín M, Close T, Costa AF, Benko-Iseppon AM, Pedrosa-Harand A, Brasileiro-Vidal AC (2020) Breaks of macrosynteny and collinearity among moth bean (Vigna aconitifolia), cowpea (V. unguiculata), and common bean (Phaseolus vulgaris). Chromosom Res. https://doi.org/10.1007/s10577-020-09635-0
Qu M, Li K, Han Y, Chen L, Li Z, Han Y (2017) Integrated karyotyping of woodland strawberry (Fragaria vesca) with oligopaint FISH probes. Cytogenet Genome Res 153:158–164. https://doi.org/10.1159/000485283
Raskina O, Barber JC, Nevo E, Belyayev A (2008) Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes. Cytogenet Genome Res 120:351–357. https://doi.org/10.1159/000121084
Ribeiro T, Vasconcelos E, Santos KGB, Vaio M, Brasileiro-Vidal AC, Pedrosa-Harand A (2019) Diversity of repetitive sequences within compact genomes of Phaseolus L. beans and allied genera Cajanus L. and Vigna Savi. Chromosom Res 28:139–153. https://doi.org/10.1007/s10577-019-09618-w
Sakai H, Naito K, Ogiso-Tanaka E, Takahashi Y, Iseki K, Muto C, Satou K, Teruya K, Shiroma A, Shimoji M, Hirano T, Itoh T, Kaga A, Tomooka N (2015) The power of single molecule real-time sequencing technology in the de novo assembly of a eukaryotic genome. Sci Rep 5:16780. https://doi.org/10.1038/srep16780
Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, Fujishiro T, Katoh M, Kohara M, Kishida Y, Minami C, Nakayama S, Nakazaki N, Shimizu Y, Shinpo S, Takahashi C, Wada T, Yamada M, Ohmido N, Hayashi M, Fukui K, Baba T, Nakamichi T, Mori H, Tabata S (2008) Genome structure of the legume, Lotus japonicus. DNA Res 15:227–239. https://doi.org/10.1093/dnares/dsn008
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183. https://doi.org/10.1038/nature08670
Schmutz J, McClean PE, Mamidi S et al (2014) A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet 46:707–713. https://doi.org/10.1038/ng.3008
Schubert I (2018) What is behind “centromere repositioning”? Chromosoma 127:229–234. https://doi.org/10.1007/s00412-018-0672-y
Šimoníková D, Němečková A, Karafiátová M, Uwimana B, Swennen R, Doležel J, Hřibová E (2019) Chromosome painting facilitates anchoring reference genome sequence to chromosomes in situ and integrated karyotyping in banana (Musa spp.). Front Plant Sci 10:1503. https://doi.org/10.3389/fpls.2019.01503
Song X, Song R, Zhou J, Yan W, Zhang T, Sun H, Xiao J, Wu Y, Xi M, Lou Q, Wang H, Wang X (2020) Development and application of oligonucleotide-based chromosome painting for chromosome 4D of Triticum aestivum L. Chromosom Res 28:171–182. https://doi.org/10.1007/s10577-020-09627-0
Tang H, Krishnakumar V, Bidwell S et al (2014) An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics 15:312. https://doi.org/10.1186/1471-2164-15-312
Vallejos CE, Sakiyama NS, Chase CD (1992) A molecular marker-based linkage map of Phaseolus vulgaris L. Genetics 131:733–740
Valliyodan B, Cannon SB, Bayer PE, Shu S, Brown AV, Ren L, Jenkins J, Chung CYL, Chan TF, Daum CG, Plott C, Hastie A, Baruch K, Barry KW, Huang W, Patil G, Varshney RK, Hu H, Batley J, Yuan Y, Song Q, Stupar RM, Goodstein DM, Stacey G, Lam HM, Jackson SA, Schmutz J, Grimwood J, Edwards D, Nguyen HT (2019) Construction and comparison of three reference-quality genome assemblies for soybean. Plant J 100:1066–1082. https://doi.org/10.1111/tpj.14500
Vasconcelos EV, Fonsêca AFA, Pedrosa-Harand A, Bortoleti KCA, Benko-Iseppon AM, da Costa AF, Brasileiro-Vidal AC (2015) Intra- and interchromosomal rearrangements between cowpea [Vigna unguiculata (L.) Walp.] and common bean (Phaseolus vulgaris L.) revealed by BAC-FISH. Chromosom Res 23:253–266. https://doi.org/10.1007/s10577-014-9464-2
Wang L, Kikuchi S, Muto C, Naito K, Isemura T, Ishimoto M, Cheng X, Kaga A, Tomooka N (2015) Reciprocal translocation identified in Vigna angularis dominates the wild population in East Japan. J Plant Res 128:653–663. https://doi.org/10.1007/s10265-015-0720-0
Wanzenböck EM, Schöfer C, Schweizer D, Bachmair A (1997) Ribosomal transcription units integrated via T-DNA transformation associate with the nucleolus and do not require upstream repeat sequences for activity in Arabidopsis thaliana. Plant J 11:1007–1016. https://doi.org/10.1046/j.1365-313x.1997.11051007.x
Wellenreuther M, Bernatchez L (2018) Eco-evolutionary genomics of chromosomal inversions. Trends Ecol Evol 33:427–440. https://doi.org/10.1016/j.tree.2018.04.002
Xin H, Zhang T, Han Y, Wu Y, Shi J, Xi M, Jiang J (2018) Chromosome painting and comparative physical mapping of the sex chromosomes in Populus tomentosa and Populus deltoides. Chromosoma 127:313–321. https://doi.org/10.1007/s00412-018-0664-y
Xin H, Zhang T, Wu Y, Zhang W, Zhang P, Xi M, Jiang J (2019) An extraordinarily stable karyotype of the woody Populus species revealed by chromosome painting. Plant J 101:253–264. https://doi.org/10.1111/tpj.14536
Young ND, Debellé F, Oldroyd GE et al (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480:520–524. https://doi.org/10.1038/nature10625
Zhao H, Zeng Z, Koo DH, Gill BS, Birchler JA, Jiang J (2017) Recurrent establishment of de novo centromeres in the pericentromeric region of maize chromosome 3. Chromosom Res 25:299–311. https://doi.org/10.1007/s10577-017-9564-x
Zhao Q, Wang Y, Bi Y, Zhai Y, Yu X, Cheng C, Wang P, Li J, Lou Q, Chen J (2019) Oligo-painting and GISH reveal meiotic chromosome biases and increased meiotic stability in synthetic allotetraploid Cucumis ×hytivus with dysploid parental karyotypes. BMC Plant Biol 19:471. https://doi.org/10.1186/s12870-019-2060-z
Acknowledgements
We thank Embrapa Arroz e Feijão, Embrapa Meio-Norte, and IPK for providing the seeds. We also thank Timothy Close (University of California, Riverside) for providing the V. unguiculata BAC clones and Claudio César Montenegro Júnior for positioning the BAC H074C16 on the sequence map. We thank CNPq, CAPES, and FACEPE for fellowships and financial support.
Funding
This research was supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) (Grant nos. 442019/2019-0, 421968/2018-4, 433931/2018-3, 310804/2017-5, 310871/2014-0, and 313527/2017-2) and FACEPE (Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco) APQ-0409-2.02/16. The doctorate scholarship and a doctorate-abroad scholarship were provided by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), Finance Code 001 and Project no. 88881.189152/2018-01, respectively.
Author information
Authors and Affiliations
Contributions
LVM performed the BAC- and oligo-FISH experiments and wrote the paper. FOB and ARSO helped to perform the experiments. AFC performed seed multiplication. QL analyzed the sequence synteny data. HZ designed the oligo-FISH probes. APH and FOB conceived the oligo probes. MMA and TC maintained and provided V. unguiculata BAC clones. JJ provided the resources for the oligo-FISH experiments. APH, LLF, AMBI, and JJ planed the experiments and discussed the results. ACBV was doctorate supervisor of LVM, designed and directed the research, and helped to write the paper. All authors read, discussed, and approved the final version of the paper.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Fig. S1
Detailed sequence synteny between Vigna angularis (Va) and V. unguiculata (Vu) pseudomolecules 2 using V. unguiculata sequences but colored in accordance with Pv2 (green) and Pv3 (red). The pericentric inversion, previously identified by our BAC-FISH analysis, was confirmed by our sequence analysis: H074C16 Vu BAC is located at 5.6 Mb and 18.7 Mb region of Va2 and Vu2, respectively, that corresponds to a red region in both species. Lateral black squares on Va2 and Vu2 represent the centromere positions. Bar =5 Mb (PNG 808 kb).
Rights and permissions
About this article
Cite this article
do Vale Martins, L., de Oliveira Bustamante, F., da Silva Oliveira, A.R. et al. BAC- and oligo-FISH mapping reveals chromosome evolution among Vigna angularis, V. unguiculata, and Phaseolus vulgaris. Chromosoma 130, 133–147 (2021). https://doi.org/10.1007/s00412-021-00758-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-021-00758-9