Abstract
Cognitive impairment is a common feature in schizophrenia and the strongest prognostic factor for long-term outcome. Identifying a trait associated with the genetic background for cognitive outcome in schizophrenia may aid in a deeper understanding of clinical disease subtypes. Fast sleep spindles may represent such a biomarker as they are strongly genetically determined, associated with cognitive functioning and impaired in schizophrenia and unaffected relatives. We measured fast sleep spindle density in 150 healthy adults and investigated its association with a genome-wide polygenic score for schizophrenia (SCZ-PGS). The association between SCZ-PGS and fast spindle density was further characterized by stratifying it to the genetic background of intelligence. SCZ-PGS was positively associated with fast spindle density. This association mainly depended on pro-cognitive genetic variants. Our results strengthen the evidence for a genetic background of spindle abnormalities in schizophrenia. Spindle density might represent an easily accessible marker for a favourable cognitive outcome which should be further investigated in clinical samples.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Schizophrenia is one of the most debilitating psychiatric disorders and has a strong genetic background [1]. It is a highly polygenic disorder, i.e., a large number of common variants contribute each with a small effect to the disorder risk [1]. Recent genome-wide association studies (GWAS) for schizophrenia identified genetic variation in more than 100 loci being associated with schizophrenia [2,3,4].
Cognitive impairment is a common feature in schizophrenia and has been reported as the strongest prognostic factor for long-term outcome [5]. GWAS reveal extensive genetic overlap between schizophrenia and intelligence [6, 7] with the direction of the majority of overlapping loci representing a higher risk for schizophrenia and poorer cognitive performance [6]. Schizophrenia is a highly heterogeneous disorder with variation in cognitive functioning that is partly explained by genetic factors [8, 9]. Recently, genetic variance associated with educational attainment—often used in large studies as a proxy for intelligence/cognition—has been shown to aid in dissecting genetic heterogeneity of schizophrenia [10]. The authors found strong genetic overlap between educational attainment and schizophrenia, with associated variants showing both concordant and discordant effects, and the results indicating at least two disease subtypes of schizophrenia, one related to high educational attainment and resembling bipolar disorder, and the other closer to a cognitive disorder, independent of bipolar disorder [10]. In an independent sample of patients with schizophrenia, genetic risk variants for schizophrenia concordant for educational attainment showed significant associations with global assessment of functioning and negative symptoms not detected using all schizophrenia risk variants or the ones discordant with educational attainment [10]. Thus, identifying a subset of variants associated with schizophrenia and a concordant effect on cognitive function may aid in characterizing clinically differing disease subtypes. In this perspective, the fast sleep spindle phenotype may be helpful for differentiating between these subtypes, especially taking into account its strong genetic background [11,12,13], its association with genetic risk for schizophrenia [14,15,16] and ample evidence for an association with intellectual capacities [17,18,19,20,21,22].
Sleep spindles are brief bursts of oscillatory EEG activity within the sigma frequency range of NREM sleep [23]. They are a defining feature of stage 2 sleep [24] and also occur in slow wave sleep. Studies investigating the heritability of sleep spindles indicate a substantial genetic background [11,12,13]. A study characterizing sleep spindles in 415 related subjects estimated spindle density to have a heritability of ~ 0.45 [12]. Also, the heritability of the NREM sleep EEG frequency band between 8 and 15.75 Hz containing sleep spindles has been estimated to be up to 0.96 in a twin study [11]. Two types of sleep spindles can be distinguished depending on their frequency (9–12 Hz slow spindles, 12–15 Hz fast spindles) and their recruitment of partially segregated cortical networks [23, 25, 26]. It has been shown that spindles play an important role in cognitive processes as they facilitate synaptic plasticity [27] and correlate with intellectual capacities [17,18,19,20,21,22] and with memory consolidation in healthy subjects [19, 28,29,30,31], patients with schizophrenia [32,33,34], and healthy first-degree relatives (FDR) of patients with schizophrenia [15]. Recently, genetic variation at ZNF804A, which is robustly associated with schizophrenia, has been correlated with altered cognitive task-dependent dynamics of sleep oscillations including spindles [35]. The neuroanatomical substrate of spindle generation is the thalamo-cortical network [36], which is implicated in functional abnormalities in schizophrenia [37,38,39]. In patients with chronic and with first-episode schizophrenia as well as in healthy FDR fast sleep spindle density has repeatedly been shown to be reduced [15, 16, 32, 33, 40,41,42,43,44,45,46]. Most data on the sleep spindle deficit in schizophrenia refer to fast spindles. In a study investigating both spindle types in schizophrenia and FDR slow spindles were spared from the spindle deficit [15].
Polygenic score (PGS) analyses represent a powerful tool to investigate the association of the genetic predisposition for a disorder or trait with other phenotypes of interest. PGS estimation uses GWAS results to predict genetic risk for each individual in an independent genotyped sample, by multiplying the allele count of each genotyped variant with its effect size in the respective GWAS [47]. Recently, Merikanto and colleagues investigated whether the polygenic risk score for schizophrenia (SCZ-PGS) based on the GWAS by Ripke et al. [2] was associated with sleep spindle activity in a sample of 150 healthy adolescents [14].
Their findings showed fast spindle density and amplitude in the central derivation to be positively correlated with schizophrenia-related genetic risk, thus in the opposite direction to all previous studies in chronic schizophrenia patients, unmedicated first-episode patients and FDR and to recent data from an animal model for schizophrenia [48]. This raises the question of how to explain the apparently contradictory findings with respect to the direction of the association between spindle activity and genetic risk for schizophrenia in healthy [14] versus clinical samples [15, 16, 32, 33, 40,41,42,43,44,45,46]. In view of the significant genetic overlap between schizophrenia and cognition [6, 7] and the strong association between spindle activity and cognitive function [17,18,19,20,21,22], stratification of the genetic risk for schizophrenia into concordant and discordant cognitive genetic variants may help to elucidate the nature of the association of the spindle phenotype with the genetic background of schizophrenia.
In the present study, we aimed (I) to analyze the association of the fast spindle density phenotype with the polygenic risk for schizophrenia in a sample of 150 healthy subjects and (II) to assess the role of the genetic basis of intelligence for this association by separately investigating the subset of single-nucleotide polymorphisms (SNPs) with concordant effects for schizophrenia and intelligence [49] and the subset of discordant SNPs. As the Merikanto study [14] reported an association of slow spindle activity with genetic variability in CACNA1I, a schizophrenia risk gene known to be implicated in spindle generation [50,51,52], we exploratively additionally aimed to replicate this finding.
Methods
Procedures
Participants had two nights of polysomnography in the sleep laboratory of the Central Institute of Mental Health (CIMH), Mannheim, Germany. Data of the second night were used for analysis, whereas the first night served as an adaptation night and for the exclusion of previously undiagnosed sleep disorders.
Study participants
Inclusion criteria were age 18–60 years, good subjective sleep quality, absence of any pre-diagnosed sleep disorder or neurological or psychiatric disorder. Exclusion criteria comprised sleep curtailment prior to the study, severe health problems, history of substance abuse, current medication intake and circadian abnormalities including shift work, recent travels across time zones, or advanced or delayed sleep–wake rhythms. Participants received monetary compensation for participation in the study.
The study sample comprised 150 healthy subjects of European ancestry (93 women; age 30.9 ± 11.6 years).
Polysomnographic sleep analysis
Polysomnography was performed using a Schwarzer Comlab 32 polysomnograph (Schwarzer GmbH, Munich, Germany) with standard montage according to the criteria of the American Academy of Sleep Medicine [24]. In addition to electroencephalography (EEG) in six derivations (F4–A1, C4–A1, O2–A1, F3–A2, C3–A2 and O1–A2) this included bilateral electrooculography, chin electromyography, surface electromyography of bilateral tibialis anterior muscles, ECG recording and recording of respiratory variables. EEG sampling rate was 256 Hz. Sleep stage scoring and detection of arousals were performed visually according to the criteria of the American Academy of Sleep Medicine [24]. Polysomnographic sleep characteristics are given in Table 1.
Sleep spindle detection
Fast and slow sleep spindles were analyzed during NREM sleep stages N2 and N3 using the C3-A2 derivation. The 30-s sleep epochs had to be free of artifacts and arousals in order to be included. Artifacts and arousals were identified by visual inspection. Discrete spindle events were automatically detected using a custom-made software tool (SpindleToolbox, version 3) using MATLAB R2009b (The MathWorks, Natick, Massachusetts) based on an algorithm adopted from previous studies [53, 54] and described in Schilling et al. [55]. In brief, power spectra of each participant were calculated, enabling the user to visually detect each individual’s spindle peak for fast and slow spindles. The signal was then band-pass filtered in the range ± 1.5 Hz around the detected spindle peak, and the root mean square (RMS) was calculated for each 200-ms interval of the filtered signal. A spindle was detected if the RMS signal exceeded a threshold of 1.5 standard deviations of the filtered signal for the duration of 0.5–3 s. Cases in which no slow spindle peak was visually detectable in the power spectrum were excluded from slow spindle analysis (10 cases), resulting in a sample of 140 subjects for slow spindle analysis. Fast spindle peaks were detectable in all 150 subjects. Spindle density was defined as the number of sleep spindles detected per sleep epoch of 30 s of all NREM 2 and 3 sleep epochs free of artifacts and was used as the primary quantitative measure of spindle activity. Morphological characteristics of individual spindle events (spindle amplitude and spindle duration) were additionally analyzed in an exploratory way: Spindle duration was defined as the interval between the threshold crossing points of each spindle. Spindle amplitude was calculated as the maximal spindle voltage after band-pass filtering. Spindle parameters are given in Table1.
Genotyping and quality control
DNA was extracted from whole blood. Genome-wide genotyping was performed using Global Screening Array 1.0 (Illumina, Inc., San Diego, CA, USA) at the Life&Brain facilities, Bonn, Germany. Quality control and filtering was performed using PLINK 1.9 [56] and R statistical environment, version 3.5.1, removing participants with > 0.02 missingness, heterozygosity rate >|.2|, and sex-mismatch. SNPs with a minor allele frequency of < 0.01, deviating from Hardy–Weinberg equilibrium (HWE) with a p value of < 10–6 and missing data > 0.02 were removed. Relatedness and population structure were filtered based on a SNP-set filtered for high quality (HWE p > 0.02, MAF > 0.20, missingness = 0), and LD pruning (pairwise r2 < 0.1 within a 200 SNP window). If subjects were cryptically related (\(\hat{\pi }\) > 0.20), one subject was excluded at random. Control for population stratification was performed by generating principal components and outliers, defined as deviating more than 6 SD on one of the first 20 principal components were excluded. Quality control resulted in a data set of 140 individuals and 481,956 SNPs for genetic analyses on fast spindle activity. In eight out of these 140 individuals, no slow spindle peak was detectable, thus resulting in 132 individuals for genetic analyses on slow spindle activity.
Polygenic scores
SCZ-PGS were calculated using PRSice 2.1.6 [57] based on GWAS data from the Psychiatric Genomics Consortium (Cases: n = 36,989, Controls: n = 113,075; [2]). The summary statistics were filtered, excluding SNPs with an info score < 0.9. PGS were calculated for the default p value thresholds (PT; 0.001, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 1). The PT corresponds to the minimum p-value threshold in the discovery GWAS required for inclusion of the SNPs to be included in the calculation of the PRS, e.g., PT of 1 considers all SNPs for the calculation, PT of 0.001 considers all SNPs with a p value smaller than 0.001 in the original GWAS. Selecting PGS at different thresholds, represent a trade-off, with PGS at more liberal PTs having a higher probability to include all relevant SNPs, but also an increased amount of random noise.
To explore the contribution of the genetic basis of intelligence to fast spindle density and its association with SCZ-PGS, we first calculated PGS for a GWAS of intelligence [49] and tested its association with fast spindle density. In a second step and in orientation to analyses presented in Bansal et al. [10], we split the SNPs of the SCZ GWAS into SNPs that either had concordant effects on SCZ and IQ (i.e., SNPs associated with increased Risk for SCZ (OR > 1) and higher IQ (beta > 0)/lower risk for SCZ (OR < 1) and lower IQ (beta < 0); nSNPs = 2,823,196), and SNPs with discordant effects (i.e., SNPs associated with increased risk for SCZ (OR > 1) and lower IQ (beta < 0)/lower risk for SCZ (OR < 1) and higher IQ (beta > 0); nSNPs = 3,067,373). SCZ-IQ-concordant-PGS and SCZ-IQ-discordant-PGS were then calculated based on those filtered SNP-sets as described above using the p values and effect sizes reported for each of the included SNPs in the SCZ GWAS.
To test for the specificity of the association of SCZ with fast spindle density, we additionally calculated PGS for bipolar disorder (Cases: n = 20,352, Controls: n = 31,358) [58], and PGS for depression (Cases: n = 170,756, Controls: n = 329,443; excluding 23andMe) [59].
Explorative additional analysis of CACNA1I gene association with spindle density
In analogy to the Merikanto study [14], we exploratively generated a SCZ-PGS limited to variants in the CACNA1I gene including 20 kilo bases (kb) down and upstream (Chr 22; bp 39,946,758–40,105,740). To complement the results of the CACNA1I-based SCZ-score, we performed a gene-based association test for CACNA1I using MAGMA v1.08 [60] to assess whether fast and slow spindle density was associated with genetic variation in CACNA1I at a gene level independent of their implication in SCZ, controlling for sex, age and the first ten principle components, using the same ± 20 kb borders.
Statistical analysis
The prediction accuracy of all PGS was assessed in PRSice 2.1.6 using the R2 statistics attributable to the PGS, which was computed as the increase in the coefficient of determination (R2) when the PGS was added to the linear model predicting the respective sleep spindle characteristics including sex, age and the first ten principle components of population stratification as covariates [57].
All reported tests were two tailed with an α level of p < 0.05.
Results
Association of spindle activity with genome-wide polygenic score for schizophrenia
Fast spindle density was positively associated with the SCZ-PGS with the strongest association (incremental R2 = 5.2%, p = 0.0043) for a PT of 0.3 (Fig. 1A, Supplementary Table S1). Fast spindle duration also was positively associated with the SCZ-PGS (strongest association: PT = 0.3; incremental R2 = 2.9%, p = 0.026; Supplementary Table S2) but fast spindle amplitude was not (all p > 0.676; Supplementary Table S3).
Association of fast spindle density with polygenic scores. A Fast spindle density predicted by schizophrenia polygenic score (SCZ-PGS). B Fast spindle density predicted by intelligence polygenic score (IQ-PGS). C Fast spindle density predicted by polygenic score based on variants with concordant effects on SCZ and intelligence (SCZ-IQ-concordant-PGS). D Fast spindle density predicted by polygenic score based on variants with discordant effects on SCZ and intelligence (SCZ-IQ-discordant-PGS). Left panels: The increase in R2 by adding the PGS to the model is shown for PGS calculated at 8 selected p value thresholds. The observed p-value is depicted on top of each bar. Right panels: Change in phenotype by PGS quintile is depicted with the lowest quintile as reference. N = 140 for all panels
SCZ-PGS was not associated with slow spindle density (all p > 0.493), or slow spindle duration (all p > 0.135), and showed a nominally significant negative association with slow spindle amplitude at one PT (PT = 0.001, R2 = 0.0283, p = 0.0472; Supplementary Tables S4–6)).
Association of fast spindle density with polygenic score for intelligence (IQ)
Fast spindle density was positively associated with the PGS for intelligence with the strongest association (incremental R2 = 3.3%, p = 0.0246) for a PT of 0.2 (Fig. 1B, Supplementary Table S7).
Association of fast spindle density with polygenic score for schizophrenia: distinction between polymorphisms concordant or discordant for schizophrenia and intelligence
The SCZ-IQ-concordant-PGS was positively associated with fast spindle density (strongest association: incremental R2 = 5.5%, p = 0.00344; PT 0.5; Fig. 1C, Supplementary Table S8). The association of SCZ-IQ-discordant-PGS with fast spindles did not reach significance (strongest association: incremental R2 = 2.4%, p = 0.0576; PT 0.4; Fig. 1D, Supplementary Table S9).
Association of fast spindle density with polygenic scores for major depression and bipolar disorder
Both Depression and Bipolar-PGS did not show a significant association with fast spindle density (Strongest association: Depression: R2 = 0.51%, p = 0.377, PT = 1, Supplementary Table S10, Bipolar: R2 = 2.1%, p = 0.0717, PT = 0.5, Supplementary Table S11).
Association of spindle density with CACNA1I genetic variance
The CACNA1I-based SCZ-score (generated in analogy to the study of Merikanto [14] for explorative analysis) was neither significantly associated with fast spindle density (all p > 0.164), fast spindle amplitude (all p > 0.320), nor fast spindle duration (all p > 0.381; Supplementary Tables S12–14). Additionally, it was neither associated with slow spindle density (all p > 0.834), slow spindle amplitude (all p > 0.0891) nor slow spindle duration (all p > 0.0551; Supplementary Tables S15–S17).
However, the gene-based analysis using MAGMA revealed a nominal significant association between genetic variation in CACNA1I and fast spindle density (p = 0.0193), but not slow spindle density (p = 0.183; Supplementary Table S18). Four SNPs showed association with fast spindle density reaching nominal significance. Information on the included SNPs and their associations with slow and fast spindle density can be found in Supplementary Table S19 and Supplementary Table S20, respectively.
Discussion
To elucidate the genetic background of impaired spindle activity in schizophrenia, we investigated the association of fast sleep spindle density with the SCZ-PGS in healthy adult subjects. In view of the important role of spindle activity for intelligence and cognition [17,18,19,20,21,22] we further analyzed the effect of stratification of genetic variance for SNPs concordant and discordant for intelligence and schizophrenia-related risk on this association. We used a genome-wide SCZ-PGS derived from Ripke et al. [2] and a PGS based on a large GWAS for intelligence [49]. In further explorative analyses, we investigated whether these findings were specific to fast versus slow spindles and to schizophrenia compared to bipolar disorder and depression.
We found (1) a positive correlation of PGS for SCZ with fast spindle density and (2) that this association was predominantly driven by the subset of SNPs concordant for schizophrenia and intelligence. The positive association of fast spindle density with genetic background of the disease was specific to schizophrenia: There was no association with PGS for major depression [59] and only a trend level association with PGS for bipolar disorder [58]. Also, association of SCZ-PGS with spindle density was specific to fast but not slow spindle density.
Our finding of a positive correlation of SCZ-PGS with fast spindle density is in line with recent results by Merikanto et al. [14]. In contrast, with regard to the report by Merikanto et al. [14] on an association of slow spindle parameters with a genetic risk score specific for the CACNA1I gene, a schizophrenia risk gene coding for the voltage-gated calcium channel subunit alpha 1 known to be implicated in spindle generation [50,51,52], in a sample of 150 healthy adolescents [14], we did not replicate this result in our explorative analysis in 150 adults. However, we did find preliminary evidence for an association of genetic variance within the CACNA1I region with fast spindle density on a gene level. This will have to be further investigated in larger samples.
Our data of a correlation of SCZ-PGS with fast spindle density strengthen the evidence for a genetic background of spindle abnormalities in schizophrenia. They further indicate that the strongest effects of genetic SCZ risk can be observed in fast spindles. Correspondingly, previous data in schizophrenia investigating both spindle types found a fast but not slow spindle deficit [15].
Importantly, both our findings and the findings of the Merikanto study are unexpected with respect to the direction of the correlation of genetic risk for schizophrenia with spindle density as data on spindle abnormalities in patients with schizophrenia consistently report on spindle deficits instead of an increase [15, 16, 32, 33, 40,41,42,43,44,45,46, 61]. Findings of opposing direction in healthy subjects compared to patient samples cannot be attributed to the effect of chronic disease course or medication in the patients, as deficient spindle activity is also reported in healthy first-degree relatives of patients with schizophrenia [15, 16, 62] and in untreated first-episode patients [16, 44]. This challenges the current straightforward concept of a genetically mediated spindle deficit in schizophrenia.
In view of the recent genetic evidence for a strong role of cognitive function (with educational attainment as a proxy) for identifying genetic heterogeneity within schizophrenia [10] and the important role of spindle activity for cognition [17,18,19,20,21,22], we analyzed the relation of the correlation between fast spindle density and schizophrenia-PGS to genome-wide genetic variability associated with intelligence (IQ-PGS) [49]. As expected fast spindle density in healthy subjects correlated with the IQ-PGS (which also is a novel finding). Distinction of two subsets of SNPs depending on whether they show concordance or discordance between intelligence and schizophrenia revealed that the positive association of SCZ-PGS with fast sleep spindle density in healthy subjects is mainly driven by polymorphisms concordant for schizophrenia and intelligence. We hypothesize that in healthy subjects the positive association between fast spindle density and SCZ-PGS mainly represents a pro-cognitive part within the spectrum of genetic risk for schizophrenia. As according to Bansal et al. [10] the genetic background of better cognitive function maps to a prognostically more favorable subtype of schizophrenia [10] and the association between high fast spindle density and SCZ-PGS in our study mainly depends on concordant cognitive SNPs (Fig. 1c, see also model in Fig. 2b), intact fast spindle density may be related to a subtype of schizophrenia characterized by genetically determined better cognitive functioning and prognosis. Richard et al. recently showed that cognition in schizophrenia cases was more strongly associated with PGS that index cognitive traits in the general population than with PGS for neuropsychiatric disorders [8]. In line with this, fast spindle density being associated with favourable cognitive traits in the general population [17,18,19,20,21,22] may be associated with good cognitive function within the clinical spectrum of schizophrenia. This should be further investigated in deeply phenotyped clinical samples of patients with schizophrenia.
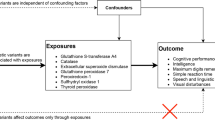
Schematic model of the postulated relationship between fast spindle density and polygenetic risk for schizophrenia (working hypothesis). Yellow areas: refer to data from this study. A Model that reconciles the data on fast sleep spindle density in healthy control samples (HC), healthy first-degree relatives (FDR) and patients with schizophrenia (SCZ); dashed line: positive correlation between schizophrenia-PGS and fast spindle density in healthy subjects (first part of this study and [14]); dotted lines: spindle deficit in patient samples and FDR compared to HC (reflecting literature data [15, 46]). B Model visualizing the postulated relationship between fast spindle density and SCZ-PGS separately for pro-cognitive variants (SNPs that are concordant for SCZ risk and intelligence; blue curve and blue dashed line) and contra-cognitive variants (SNPs that are discordant for SCZ risk and intelligence; red curve and red dashed line): The positive correlation between spindle density and SCZ-PGS mainly depends on pro-cognitive variants
As patients with schizophrenia as well as healthy FDR (both having an increased genetic load of schizophrenia-related risk alleles compared to healthy subject samples) show reduced fast spindle density, it could be speculated that the fast spindle density phenotype may follow a converted U-shaped curve with increasing number of schizophrenia risk alleles (Fig. 2a). With regard to an evolutionary framework for a deleterious disorder such as schizophrenia, it previously has been proposed that in healthy subjects inheritance of risk alleles insufficient in number to manifest as schizophrenia may manifest as a behavioral phenotype with adaptive advantages [63] (here high spindle density, high cognitive functioning). A similar relationship has been found for creativity and schizophrenia sharing genetic roots [64]. Possibly, deficient fast spindle density found in patient samples and FDR may be a correlate of a critical number and/or set of risk alleles including their interactions predisposing to actual illness. For direct evidence supporting this association, spindle density and SCZ-PGS will have to be investigated in samples of FDR and unmedicated patients, necessitating larger sample sizes than those of previous investigations of spindle activity in SCZ and FDR [46]. Also, neurobiological pathways mediating the decline of fast spindle density with increasing genetic load for schizophrenia will be interesting to investigate in view of a better understanding of disease mechanisms.
Strengths and limitations
The good characterization of our sample with exclusion of any sleep or general pathology that could have an influence on the spindle phenotype as well as the study under two-night sleep laboratory conditions are strengths of this work. Although the sample size is rather small for genetic studies, it is the same as in the study by Merikanto and colleagues on this topic [14]. We consider it an indication for the robustness of the present results, that we replicate the main finding of the study by Merikanto et al. [14], the positive association of SCZ-PGS with fast spindle density. Furthermore, the well-defined phenotype of spindle activity with its very strong genetic background allows meaningful results despite the limited sample size. The heterogeneity of algorithms used in spindle research is a potential limitation to the comparability of results. The algorithm used in our study operates with individual spindle peak identification (in contrast to fixed-frequency methods) and, thus, is advantageous for detection of fast and slow spindles taking individual differences in spindle peak-frequency into account [65]. However, future studies should aim to replicate and extend the present results.
Conclusions
Our results strengthen the evidence for a genetic background of spindle abnormalities in schizophrenia, and fit the considerations about genetically driven cognitive heterogeneity of schizophrenia [8, 10]. In this context, integrity of spindle activity might represent an easily accessible biological marker for a favorable cognitive outcome in schizophrenia. This should stimulate further research on the prognostic value of sleep spindle parameters for clinical subtypes of schizophrenia. Also our results reconcile the previous contradictory findings with regard to the direction of the association of fast spindle density with genetic background for schizophrenia (Fig. 2A, B): As now shown in two studies in independent samples (Merikanto et al. [14] and our present data), healthy subjects with lower polygenic risk for SCZ compared to patient samples and FDR, show a positive correlation between fast spindle density and polygenic risk for SCZ (Fig. 2A), whereby higher fast spindle density mainly maps to a pro-cognitive subset of genetic risk variants (Fig. 2B). Increasing genetic risk for schizophrenia in samples of FDR and patients may then potentially be associated with decreasing fast spindle density resulting in the spindle deficit repeatedly found in clinical samples [32, 33, 40,41,42,43,44,45,46] and FDR [15, 16] (Fig. 2A). Within this decline in fast spindle density from healthy subjects to clinical samples, those patients with a genetic background enriched for SNPs concordant for intelligence and SCZ related risk will be expected to show higher spindle density compared to those patients with a genetic background enriched for discordant SNPs (see working hypothesis model in Fig. 2B). This as well as the relation of spindle density with cognitive functioning and prognosis should be further investigated in clinically defined and deeply phenotyped patient samples.
References
Legge SE, Santoro ML, Periyasamy S, Okewole A, Arsalan A, Kowalec K (2021) Genetic architecture of schizophrenia: a review of major advancements. Psychol Med 51:1–10
Schizophrenia Working Group of the Psychiatric Genomics C (2014) Biological insights from 108 schizophrenia-associated genetic loci. Nature 511:421–427
Pardinas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, Legge SE, Bishop S, Cameron D, Hamshere ML, Han J, Hubbard L, Lynham A, Mantripragada K, Rees E, MacCabe JH, McCarroll SA, Baune BT, Breen G, Byrne EM, Dannlowski U, Eley TC, Hayward C, Martin NG, McIntosh AM, Plomin R, Porteous DJ, Wray NR, Caballero A, Geschwind DH, Huckins LM, Ruderfer DM, Santiago E, Sklar P, Stahl EA, Won H, Agerbo E, Als TD, Andreassen OA, Baekvad-Hansen M, Mortensen PB, Pedersen CB, Borglum AD, Bybjerg-Grauholm J, Djurovic S, Durmishi N, Pedersen MG, Golimbet V, Grove J, Hougaard DM, Mattheisen M, Molden E, Mors O, Nordentoft M, Pejovic-Milovancevic M, Sigurdsson E, Silagadze T, Hansen CS, Stefansson K, Stefansson H, Steinberg S, Tosato S, Werge T, Consortium G, Consortium C, Collier DA, Rujescu D, Kirov G, Owen MJ, O’Donovan MC, Walters JTR (2018) Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet 50:381–389
Lam M, Chen CY, Li Z, Martin AR, Bryois J, Ma X, Gaspar H, Ikeda M, Benyamin B, Brown BC, Liu R, Zhou W, Guan L, Kamatani Y, Kim SW, Kubo M, Kusumawardhani A, Liu CM, Ma H, Periyasamy S, Takahashi A, Xu Z, Yu H, Zhu F, Schizophrenia Working Group of the Psychiatric Genomics C, Indonesia Schizophrenia C, Genetic Rosn-C, the N, Chen WJ, Faraone S, Glatt SJ, He L, Hyman SE, Hwu HG, McCarroll SA, Neale BM, Sklar P, Wildenauer DB, Yu X, Zhang D, Mowry BJ, Lee J, Holmans P, Xu S, Sullivan PF, Ripke S, O’Donovan MC, Daly MJ, Qin S, Sham P, Iwata N, Hong KS, Schwab SG, Yue W, Tsuang M, Liu J, Ma X, Kahn RS, Shi Y, Huang H (2019) Comparative genetic architectures of schizophrenia in east Asian and European populations. Nat Genet 51:1670–1678
Green MF, Kern RS, Braff DL, Mintz J (2000) Neurocognitive deficits and functional outcome in schizophrenia: are we measuring the “right stuff”? Schizophr Bull 26:119–136
Smeland OB, Bahrami S, Frei O, Shadrin A, O’Connell K, Savage J, Watanabe K, Krull F, Bettella F, Steen NE, Ueland T, Posthuma D, Djurovic S, Dale AM, Andreassen OA (2020) Genome-wide analysis reveals extensive genetic overlap between schizophrenia, bipolar disorder, and intelligence. Mol Psychiatry 25:844–853
Hubbard L, Tansey KE, Rai D, Jones P, Ripke S, Chambert KD, Moran JL, McCarroll SA, Linden DE, Owen MJ, O’Donovan MC, Walters JT, Zammit S (2016) Evidence of common genetic overlap between schizophrenia and cognition. Schizophr Bull 42:832–842
Richards AL, Pardiñas AF, Frizzati A, Tansey KE, Lynham AJ, Holmans P, Legge SE, Savage JE, Agartz I, Andreassen OA, Blokland GAM, Corvin A, Cosgrove D, Degenhardt F, Djurovic S, Espeseth T, Ferraro L, Gayer-Anderson C, Giegling I, van Haren NE, Hartmann AM, Hubert JJ, Jönsson EG, Konte B, Lennertz L, Olde Loohuis LM, Melle I, Morgan C, Morris DW, Murray RM, Nyman H, Ophoff RA, Investigators G, van Os J, Group EW, Schizophrenia Working Group of the Psychiatric Genomics C, Petryshen TL, Quattrone D, Rietschel M, Rujescu D, Rutten BPF, Streit F, Strohmaier J, Sullivan PF, Sundet K, Wagner M, Escott-Price V, Owen MJ, Donohoe G, O’Donovan MC, Walters JTR (2020) The relationship between polygenic risk scores and cognition in schizophrenia. Schizophr Bull 46:336–344
Lam M, Hill WD, Trampush JW, Yu J, Knowles E, Davies G, Stahl E, Huckins L, Liewald DC, Djurovic S, Melle I, Sundet K, Christoforou A, Reinvang I, DeRosse P, Lundervold AJ, Steen VM, Espeseth T, Raikkonen K, Widen E, Palotie A, Eriksson JG, Giegling I, Konte B, Hartmann AM, Roussos P, Giakoumaki S, Burdick KE, Payton A, Ollier W, Chiba-Falek O, Attix DK, Need AC, Cirulli ET, Voineskos AN, Stefanis NC, Avramopoulos D, Hatzimanolis A, Arking DE, Smyrnis N, Bilder RM, Freimer NA, Cannon TD, London E, Poldrack RA, Sabb FW, Congdon E, Conley ED, Scult MA, Dickinson D, Straub RE, Donohoe G, Morris D, Corvin A, Gill M, Hariri AR, Weinberger DR, Pendleton N, Bitsios P, Rujescu D, Lahti J, Le Hellard S, Keller MC, Andreassen OA, Deary IJ, Glahn DC, Malhotra AK, Lencz T (2019) Pleiotropic meta-analysis of cognition, education, and schizophrenia differentiates roles of early neurodevelopmental and adult synaptic pathways. Am J Hum Genet 105:334–350
Bansal V, Mitjans M, Burik CAP, Linner RK, Okbay A, Rietveld CA, Begemann M, Bonn S, Ripke S, de Vlaming R, Nivard MG, Ehrenreich H, Koellinger PD (2018) Genome-wide association study results for educational attainment aid in identifying genetic heterogeneity of schizophrenia. Nat Commun 9:3078
De Gennaro L, Marzano C, Fratello F, Moroni F, Pellicciari MC, Ferlazzo F, Costa S, Couyoumdjian A, Curcio G, Sforza E, Malafosse A, Finelli LA, Pasqualetti P, Ferrara M, Bertini M, Rossini PM (2008) The electroencephalographic fingerprint of sleep is genetically determined: A twin study. Ann Neurol 64:455–460
Purcell SM, Manoach DS, Demanuele C, Cade BE, Mariani S, Cox R, Panagiotaropoulou G, Saxena R, Pan JQ, Smoller JW, Redline S, Stickgold R (2017) Characterizing sleep spindles in 11,630 individuals from the national sleep research resource. Nat Commun 8:15930
Ambrosius U, Lietzenmaier S, Wehrle R, Wichniak A, Kalus S, Winkelmann J, Bettecken T, Holsboer F, Yassouridis A, Friess E (2008) Heritability of sleep electroencephalogram. Biol Psychiat 64:344–348
Merikanto I, Utge S, Lahti J, Kuula L, Makkonen T, Lahti-Pulkkinen M, Heinonen K, Raikkonen K, Andersson S, Strandberg T, Pesonen AK (2019) Genetic risk factors for schizophrenia associate with sleep spindle activity in healthy adolescents. J Sleep Res 28:e12762
Schilling C, Schlipf M, Spietzack S, Rausch F, Eisenacher S, Englisch S, Reinhard I, Haller L, Grimm O, Deuschle M, Tost H, Zink M, Meyer-Lindenberg A, Schredl M (2017) Fast sleep spindle reduction in schizophrenia and healthy first-degree relatives: association with impaired cognitive function and potential intermediate phenotype. Eur Arch Psychiatry Clin Neurosci 267:213–224
Manoach DS, Demanuele C, Wamsley EJ, Vangel M, Montrose DM, Miewald J, Kupfer D, Buysse D, Stickgold R, Keshavan MS (2014) Sleep spindle deficits in antipsychotic-naive early course schizophrenia and in non-psychotic first-degree relatives. Front Hum Neurosci 8:762
Bodizs R, Kis T, Lazar AS, Havran L, Rigo P, Clemens Z, Halasz P (2005) Prediction of general mental ability based on neural oscillation measures of sleep. J Sleep Res 14:285–292
Schabus M, Hodlmoser K, Gruber G, Sauter C, Anderer P, Klosch G, Parapatics S, Saletu B, Klimesch W, Zeitlhofer J (2006) Sleep spindle-related activity in the human eeg and its relation to general cognitive and learning abilities. Eur J Neurosci 23:1738–1746
Fogel SM, Smith CT (2011) The function of the sleep spindle: a physiological index of intelligence and a mechanism for sleep-dependent memory consolidation. Neurosci Biobehav Rev 35:1154–1165
Ujma PP, Konrad BN, Genzel L, Bleifuss A, Simor P, Potari A, Kormendi J, Gombos F, Steiger A, Bodizs R, Dresler M (2014) Sleep spindles and intelligence: evidence for a sexual dimorphism. J Neurosci 34:16358–16368
Fang Z, Sergeeva V, Ray LB, Viczko J, Owen AM, Fogel SM (2017) Sleep spindles and intellectual ability: epiphenomenon or directly related? J Cogn Neurosci 29:167–182
Fogel SM, Nader R, Cote KA, Smith CT (2007) Sleep spindles and learning potential. Behav Neurosci 121:1–10
De Gennaro L, Ferrara M (2003) Sleep spindles: an overview. Sleep Med Rev 7:423–440
Iber C, Ancoli-Israel S, Chesson A, Quan SFftAAoSM (2007) The aasm manual for the scoring of sleep and associated events: rules, terminology, and technical specifications. American Academy of Sleep Medicine, Westchester
Anderer P, Klosch G, Gruber G, Trenker E, Pascual-Marqui RD, Zeitlhofer J, Barbanoj MJ, Rappelsberger P, Saletu B (2001) Low-resolution brain electromagnetic tomography revealed simultaneously active frontal and parietal sleep spindle sources in the human cortex. Neuroscience 103:581–592
Schabus M, Dang-Vu TT, Albouy G, Balteau E, Boly M, Carrier J, Darsaud A, Degueldre C, Desseilles M, Gais S, Phillips C, Rauchs G, Schnakers C, Sterpenich V, Vandewalle G, Luxen A, Maquet P (2007) Hemodynamic cerebral correlates of sleep spindles during human non-rapid eye movement sleep. Proc Natl Acad Sci USA 104:13164–13169
Rosanova M, Ulrich D (2005) Pattern-specific associative long-term potentiation induced by a sleep spindle-related spike train. J Neurosci 25:9398–9405
Clemens Z, Fabo D, Halasz P (2005) Overnight verbal memory retention correlates with the number of sleep spindles. Neuroscience 132:529–535
Schabus M, Gruber G, Parapatics S, Sauter C, Klosch G, Anderer P, Klimesch W, Saletu B, Zeitlhofer J (2004) Sleep spindles and their significance for declarative memory consolidation. Sleep 27:1479–1485
Tamaki M, Matsuoka T, Nittono H, Hori T (2008) Fast sleep spindle (13–15 hz) activity correlates with sleep-dependent improvement in visuomotor performance. Sleep 31:204–211
Rasch B, Pommer J, Diekelmann S, Born J (2009) Pharmacological rem sleep suppression paradoxically improves rather than impairs skill memory. Nat Neurosci 12:396–397
Goder R, Graf A, Ballhausen F, Weinhold S, Baier PC, Junghanns K, Prehn-Kristensen A (2015) Impairment of sleep-related memory consolidation in schizophrenia: relevance of sleep spindles? Sleep Med 16:564–569
Wamsley EJ, Tucker MA, Shinn AK, Ono KE, McKinley SK, Ely AV, Goff DC, Stickgold R, Manoach DS (2012) Reduced sleep spindles and spindle coherence in schizophrenia: mechanisms of impaired memory consolidation? Biol Psychiatry 71:154–161
Au CH, Harvey CJ (2020) Systematic review: the relationship between sleep spindle activity with cognitive functions, positive and negative symptoms in psychosis. Sleep Med X 2:100025
Bartsch U, Corbin LJ, Hellmich C, Taylor M, Easey KE, Durant C, Marston HM, Timpson NJ, Jones MW (2021) Schizophrenia-associated variation at znf804a correlates with altered experience-dependent dynamics of sleep slow waves and spindles in healthy young adults. Sleep. https://doi.org/10.1093/sleep/zsab191
Fuentealba P, Steriade M (2005) The reticular nucleus revisited: intrinsic and network properties of a thalamic pacemaker. Prog Neurobiol 75:125–141
Woodward ND, Karbasforoushan H, Heckers S (2012) Thalamocortical dysconnectivity in schizophrenia. Am J Psychiatry 169:1092–1099
Anticevic A, Cole MW, Repovs G, Murray JD, Brumbaugh MS, Winkler AM, Savic A, Krystal JH, Pearlson GD, Glahn DC (2014) Characterizing thalamo-cortical disturbances in schizophrenia and bipolar illness. Cereb Cortex 24:3116–3130
Anticevic A, Haut K, Murray JD, Repovs G, Yang GJ, Diehl C, McEwen SC, Bearden CE, Addington J, Goodyear B, Cadenhead KS, Mirzakhanian H, Cornblatt BA, Olvet D, Mathalon DH, McGlashan TH, Perkins DO, Belger A, Seidman LJ, Tsuang MT, van Erp TG, Walker EF, Hamann S, Woods SW, Qiu M, Cannon TD (2015) Association of thalamic dysconnectivity and conversion to psychosis in youth and young adults at elevated clinical risk. JAMA Psychiat 72:882–891
Seeck-Hirschner M, Baier PC, Sever S, Buschbacher A, Aldenhoff JB, Goder R (2010) Effects of daytime naps on procedural and declarative memory in patients with schizophrenia. J Psychiatr Res 44:42–47
Ferrarelli F, Huber R, Peterson MJ, Massimini M, Murphy M, Riedner BA, Watson A, Bria P, Tononi G (2007) Reduced sleep spindle activity in schizophrenia patients. Am J Psychiatry 164:483–492
Ferrarelli F, Peterson MJ, Sarasso S, Riedner BA, Murphy MJ, Benca RM, Bria P, Kalin NH, Tononi G (2010) Thalamic dysfunction in schizophrenia suggested by whole-night deficits in slow and fast spindles. Am J Psychiatry 167:1339–1348
Gerstenberg M, Furrer M, Tesler N, Franscini M, Walitza S, Huber R (2020) Reduced sleep spindle density in adolescent patients with early-onset schizophrenia compared to major depressive disorder and healthy controls. Schizophr Res 221:20–28
Yazıhan NT, Yetkin S (2020) Sleep, sleep spindles, and cognitive functions in drug-naive patients with first-episode psychosis. J Clin Sleep Med 16:2079–2087
Manoach DS, Thakkar KN, Stroynowski E, Ely A, McKinley SK, Wamsley E, Djonlagic I, Vangel MG, Goff DC, Stickgold R (2010) Reduced overnight consolidation of procedural learning in chronic medicated schizophrenia is related to specific sleep stages. J Psychiatr Res 44:112–120
Lai M, Hegde R, Kelly S, Bannai D, Lizano P, Stickgold R, Manoach DS, Keshavan M (2021) Investigating sleep spindle density and schizophrenia: a meta-analysis. Psychiatry Res 307:114265
Lewis CM, Vassos E (2020) Polygenic risk scores: from research tools to clinical instruments. Genome Med 12:44
Ang G, McKillop LE, Purple R, Blanco-Duque C, Peirson SN, Foster RG, Harrison PJ, Sprengel R, Davies KE, Oliver PL, Bannerman DM, Vyazovskiy VV (2018) Absent sleep eeg spindle activity in glua1 (gria1) knockout mice: relevance to neuropsychiatric disorders. Transl Psychiatry 8:154
Savage JE, Jansen PR, Stringer S, Watanabe K, Bryois J, de Leeuw CA, Nagel M, Awasthi S, Barr PB, Coleman JRI, Grasby KL, Hammerschlag AR, Kaminski JA, Karlsson R, Krapohl E, Lam M, Nygaard M, Reynolds CA, Trampush JW, Young H, Zabaneh D, Hagg S, Hansell NK, Karlsson IK, Linnarsson S, Montgomery GW, Munoz-Manchado AB, Quinlan EB, Schumann G, Skene NG, Webb BT, White T, Arking DE, Avramopoulos D, Bilder RM, Bitsios P, Burdick KE, Cannon TD, Chiba-Falek O, Christoforou A, Cirulli ET, Congdon E, Corvin A, Davies G, Deary IJ, DeRosse P, Dickinson D, Djurovic S, Donohoe G, Conley ED, Eriksson JG, Espeseth T, Freimer NA, Giakoumaki S, Giegling I, Gill M, Glahn DC, Hariri AR, Hatzimanolis A, Keller MC, Knowles E, Koltai D, Konte B, Lahti J, Le Hellard S, Lencz T, Liewald DC, London E, Lundervold AJ, Malhotra AK, Melle I, Morris D, Need AC, Ollier W, Palotie A, Payton A, Pendleton N, Poldrack RA, Raikkonen K, Reinvang I, Roussos P, Rujescu D, Sabb FW, Scult MA, Smeland OB, Smyrnis N, Starr JM, Steen VM, Stefanis NC, Straub RE, Sundet K, Tiemeier H, Voineskos AN, Weinberger DR, Widen E, Yu J, Abecasis G, Andreassen OA, Breen G, Christiansen L et al (2018) Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet 50:912–919
Astori S, Wimmer RD, Prosser HM, Corti C, Corsi M, Liaudet N, Volterra A, Franken P, Adelman JP, Luthi A (2011) The ca(v)3.3 calcium channel is the major sleep spindle pacemaker in thalamus. Proc Natl Acad Sci USA 108:13823–13828
Pellegrini C, Lecci S, Luthi A, Astori S (2016) Suppression of sleep spindle rhythmogenesis in mice with deletion of cav3.2 and cav3.3 t-type ca(2+) channels. Sleep 39:875–885
Ghoshal A, Uygun DS, Yang L, McNally JM, Lopez-Huerta VG, Arias-Garcia MA, Baez-Nieto D, Allen A, Fitzgerald M, Choi S, Zhang Q, Hope JM, Yan K, Mao X, Nicholson TB, Imaizumi K, Fu Z, Feng G, Brown RE, Strecker RE, Purcell SM, Pan JQ (2020) Effects of a patient-derived de novo coding alteration of cacna1i in mice connect a schizophrenia risk gene with sleep spindle deficits. Transl Psychiatry 10:29
Gais S, Molle M, Helms K, Born J (2002) Learning-dependent increases in sleep spindle density. J Neurosci 22:6830–6834
Molle M, Marshall L, Gais S, Born J (2002) Grouping of spindle activity during slow oscillations in human non-rapid eye movement sleep. J Neurosci 22:10941–10947
Schilling C, Gappa L, Schredl M, Streit F, Treutlein J, Frank J, Deuschle M, Meyer-Lindenberg A, Rietschel M, Witt SH (2018) Fast sleep spindle density is associated with rs4680 (val108/158met) genotype of catechol-o-methyltransferase (comt). Sleep 41
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ (2015) Second-generation plink: Rising to the challenge of larger and richer datasets. Gigascience 4:7
Choi SW, O'Reilly PF (2019) Prsice-2: Polygenic risk score software for biobank-scale data. Gigascience 8
Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, Mattheisen M, Wang Y, Coleman JRI, Gaspar HA, de Leeuw CA, Steinberg S, Pavlides JMW, Trzaskowski M, Byrne EM, Pers TH, Holmans PA, Richards AL, Abbott L, Agerbo E, Akil H, Albani D, Alliey-Rodriguez N, Als TD, Anjorin A, Antilla V, Awasthi S, Badner JA, Baekvad-Hansen M, Barchas JD, Bass N, Bauer M, Belliveau R, Bergen SE, Pedersen CB, Boen E, Boks MP, Boocock J, Budde M, Bunney W, Burmeister M, Bybjerg-Grauholm J, Byerley W, Casas M, Cerrato F, Cervantes P, Chambert K, Charney AW, Chen D, Churchhouse C, Clarke TK, Coryell W, Craig DW, Cruceanu C, Curtis D, Czerski PM, Dale AM, de Jong S, Degenhardt F, Del-Favero J, DePaulo JR, Djurovic S, Dobbyn AL, Dumont A, Elvsashagen T, Escott-Price V, Fan CC, Fischer SB, Flickinger M, Foroud TM, Forty L, Frank J, Fraser C, Freimer NB, Frisen L, Gade K, Gage D, Garnham J, Giambartolomei C, Pedersen MG, Goldstein J, Gordon SD, Gordon-Smith K, Green EK, Green MJ, Greenwood TA, Grove J, Guan W, Guzman-Parra J, Hamshere ML, Hautzinger M, Heilbronner U, Herms S, Hipolito M, Hoffmann P, Holland D, Huckins L, Jamain S, Johnson JS, Jureus A et al (2019) Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet 51:793–803
Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M, Coleman JRI, Hagenaars SP, Ward J, Wigmore EM, Alloza C, Shen X, Barbu MC, Xu EY, Whalley HC, Marioni RE, Porteous DJ, Davies G, Deary IJ, Hemani G, Berger K, Teismann H, Rawal R, Arolt V, Baune BT, Dannlowski U, Domschke K, Tian C, Hinds DA, and Me Research T, Major Depressive Disorder Working Group of the Psychiatric Genomics C, Trzaskowski M, Byrne EM, Ripke S, Smith DJ, Sullivan PF, Wray NR, Breen G, Lewis CM, McIntosh AM (2019) Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci 22:343–352
de Leeuw CA, Mooij JM, Heskes T, Posthuma D (2015) Magma: generalized gene-set analysis of gwas data. PLoS Comput Biol 11:e1004219
Goder R, Fritzer G, Gottwald B, Lippmann B, Seeck-Hirschner M, Serafin I, Aldenhoff JB (2008) Effects of olanzapine on slow wave sleep, sleep spindles and sleep-related memory consolidation in schizophrenia. Pharmacopsychiatry 41:92–99
D’Agostino A, Castelnovo A, Cavallotti S, Casetta C, Marcatili M, Gambini O, Canevini M, Tononi G, Riedner B, Ferrarelli F, Sarasso S (2018) Sleep endophenotypes of schizophrenia: Slow waves and sleep spindles in unaffected first-degree relatives. NPJ Schizophr 4:2
Pearlson GD, Folley BS (2008) Schizophrenia, psychiatric genetics, and darwinian psychiatry: an evolutionary framework. Schizophr Bull 34:722–733
Power RA, Steinberg S, Bjornsdottir G, Rietveld CA, Abdellaoui A, Nivard MM, Johannesson M, Galesloot TE, Hottenga JJ, Willemsen G, Cesarini D, Benjamin DJ, Magnusson PK, Ullén F, Tiemeier H, Hofman A, van Rooij FJ, Walters GB, Sigurdsson E, Thorgeirsson TE, Ingason A, Helgason A, Kong A, Kiemeney LA, Koellinger P, Boomsma DI, Gudbjartsson D, Stefansson H, Stefansson K (2015) Polygenic risk scores for schizophrenia and bipolar disorder predict creativity. Nat Neurosci 18:953–955
Ujma PP, Gombos F, Genzel L, Konrad BN, Simor P, Steiger A, Dresler M, Bódizs R (2015) A comparison of two sleep spindle detection methods based on all night averages: individually adjusted versus fixed frequencies. Front Hum Neurosci 9:52. https://doi.org/10.3389/fnhum.2015.00052
World Medical A (2013) World medical association declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA 310:2191–2194
Acknowledgements
This work was supported by the German Federal Ministry of Education and Research (BMBF) through ERA-NET NEURON, “SynSchiz—Linking synaptic dysfunction to disease mechanisms in schizophrenia—a multilevel investigation” [01EW1810 to MR], through ERA-NET NEURON “Impact of Early life MetaBolic and psychosocial strEss on susceptibility to mental Disorders; from converging epigenetic signatures to novel targets for therapeutic intervention” [01EW1904 to MR].
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the German Federal Ministry of Education and Research (BMBF) through ERA-NET NEURON, “SynSchiz—Linking synaptic dysfunction to disease mechanisms in schizophrenia—a multilevel investigation” [01EW1810 to MR], through ERA-NET NEURON “Impact of Early life MetaBolic and psychosocial strEss on susceptibility to mental Disorders; from converging epigenetic signatures to novel targets for therapeutic intervention” [01EW1904 to MR].
Author information
Authors and Affiliations
Contributions
CS, FS, SW and ES conceptualized the work. CS and MS were responsible for sleep and spindle data acquisition. FS, SW, LZ and JF were responsible for genetic data acquisition and statistical analyses. ES, AM-L, MS, MD, MR and SW contributed to data interpretation. CS and FS drafted the manuscript. All authors revised and agreed upon the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
The study was approved by the ethics committee of the Medical Faculty Mannheim, University of Heidelberg (2011-315N-MA) and accorded with the Declaration of Helsinki [66].
Consent to participate
Prior to the study all participants provided informed written consent.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Schilling, C., Zillich, L., Schredl, M. et al. Association of polygenic risk for schizophrenia with fast sleep spindle density depends on pro-cognitive variants. Eur Arch Psychiatry Clin Neurosci 272, 1193–1203 (2022). https://doi.org/10.1007/s00406-022-01435-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00406-022-01435-3