Abstract
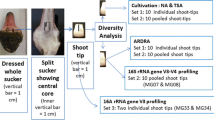
Genetic diversity of pink-pigmented facultative methylotrophic bacteria belonging to the genus, Methylobacterium, was assessed using 16S ribosomal RNA (rRNA) gene sequencing, amplified ribosomal DNA restriction analysis (ARDRA), and differential carbon-substrate utilization profile in the phyllosphere of cotton, maize, sunflower, soybean, and mentha plants. Methylobacterium populi, Methylobacterium thiocyanatum, Methylobacterium suomiense, M. aminovorans, and Methylobacterium fujisawaense were identified to colonize the phyllosphere of these crop plants. Among these, M. populi found to be the dominating species followed by M. aminovorans. The diversity indices like Shannon index of diversity, Pielou index of evenness, and Margalef index of richness calculated based on combined data of ARDRA, 16S rRNA gene sequence, and differential carbon-substrate utilization revealed that sunflower leaves showed richest methylobacterial diversity, followed by soybean, while cotton and mentha leaves recorded the lowest diversity. The outcome of this study indicates that diversity of Methylobacterium in phyllosphere depends upon the host plant species colonized.


Similar content being viewed by others
References
Abanda-Nkpwatt D, Musch M, Tschiersch J, Boettner M, Schwab W (2006) Molecular interaction between Methylobacterium extorquens and seedlings: growth promotion, methanol consumption, and localization of the methanol emission site. J Exp Bot 57:4025–4032, DOI 10.1093/jxb/erl173
Anesti V, Vohra J, Goonetilleka S, McDonald IR, Straubler B, Stackebrandt E et al (2004) Molecular detection and isolation of facultatively methylotrophic bacteria, including Methylobacterium podarium sp. nov., from the human foot microflora. Environ Microbiol 6:820–830, DOI 10.1111/j.1462-2920.2004.00623.x
Aslam Z, Lee CS, Kim K, Im W, Ten LN, Lee S (2007) Methylobacterium jeotgali sp. nov., a nonpigmented, facultatively methylotrophic bacterium isolated from jeotgal, a traditional Korean fermented seafood. Int J Syst Evol Microbiol 57:566–571, DOI 10.1099/ijs.0.64625-0
Bousfield IJ, Green PN (1985) Reclassification of bacteria of the genus Protomonas Urakami and Komagata 1984 in the genus Methylobacterium (Patt, Cole, and Hanson) emend. Green and Bousfield 1983. Int J Syst Bacteriol 35:209
Bratina BJ, Brusseau GA, Hanson RS (1992) Use of 16S rRNA analysis to investigate phylogeny of methylotrophic bacteria. Int J Syst Bacteriol 42:645–648
Brusseau GA, Bulygina ES, Hanson RS (1994) Phylogenetic analysis and development of probes for differentiating methylotrophic bacteria. Appl Environ Microbiol 60:626–636
Corpe WA, Rheem S (1989) Ecology of the methylotrophic bacteria on living leaf surfaces. FEMS Microbiol Ecol 62:243–250, DOI 10.1111/j.1574-6968.1989.tb03698.x
Doronina NV, Trotsenko YA, Tourova TP, Kuznetsov BB, Leisinger T (2000) Methylopila helvetica sp. nov. and Methylobacterium dichloromethanicum sp. nov.: novel aerobic facultatively methylotrophic bacteria utilizing dichloromethane. Syst Appl Microbiol 23:210–218
Doronina NV, Trotsenko YA, Kuznetsov BB, Tourova TP, Salkinoja-Salonen MS (2002) Methylobacterium suomiense sp. nov. and Methylobacterium lusitanum sp. nov., aerobic, pink-pigmented, facultatively methylotrophic bacteria. Int J Syst Evol Microbiol 52:773–776, DOI 10.1099/ijs.0.02014-0
Felsenstein J (1989) PHYLIP-Phylogeny inference package (version 3.2). Cladistics 5:164–166
Fournier D, Trott S, Hawari J, Spain J (2005) Metabolism of the aliphatic nitramine 4-Nitro-2,4-diazabutanal by methylobacterium sp. strain JS178. Appl Environ Microbiol 71:4199–4202, DOI 10.1128/AEM.71.8.4199-4202.2005
Gallego V, Garcia MT, Ventosa A (2005a) Methylobacterium hispanicum sp. nov. and Methylobacterium aquaticum sp. nov., isolated from drinking water. Int J Syst Evol Microbiol 55:281–287, DOI 10.1099/ijs.0.63319-0
Gallego V, Garcia MT, Ventosa A (2005b) Methylobacterium variabile sp nov, a methylotrophic bacterium isolated from an aquatic environment. Int J Syst Evol Microbiol 55:1429–1433, DOI 10.1099/ijs.0.63597-0
Gallego V, Garcia MT, Ventosa A (2005c) Methylobacterium isbiliense sp. nov., isolated from the drinking water system of Sevilla, Spain. Int J Syst Evol Microbiol 55:2333–2337, DOI 10.1099/ijs.0.63773-0
Gallego V, García MT, Ventosa A (2006) Methylobacterium adhaesivum sp. nov., a methylotrophic bacterium isolated from drinking water. Int J Syst Evol Microbiol 56:339–342, DOI 10.1099/ijs.0.63966-0
Green PN (1992) The genus Methylobacterium. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds) The prokaryotes. 2nd edn. Springer, Berlin, pp 2342–2349
Green PN, Bousfield IJ (1983) Emendation of Methylobacterium Patt, Cole, and Hanson 1976; Methylobacterium rhodinum (Heumann 1962) comb. nov. corrig.; Methylobacterium radiotolerans (Ito & Iizuka 1971) comb. nov., corrig.; and Methylobacterium mesophilicum (Austin & Goodfellow 1979) comb. nov. Int J Syst Bacteriol 33:875–877
Green PN, Bousfield IJ, Hood D (1988) Three new Methylobacterium species: M. rhodesianum sp. nov., M. zatmanii sp. nov., and M. fujisawaense sp. nov. Int J Syst Bacteriol 38:124–127
Heyndrickx M, Vauterin L, Vandamme P, Kersters K, De Vos P (1996) Applicability of combined amplified ribosomal DNA restriction analysis (ARDRA) patterns in bacterial phylogeny and taxonomy. J Microbiol Methods 26:247–259, DOI 10.1016/0167-7012(96)00916-5
Holland MA, Polacco JC (1994) PPFMs and other covert contaminants: is there more to plant physiology than just plant? Annu Rev Plant Physiol Plant Mol Biol 45:197–209, DOI 10.1146/annurev.pp.45.060194.001213
Idris R, Kuffner M, Bodrossy L, Puschenreiter M, Monchy S, Wenzel WW et al (2006) Characterization of Ni-tolerant methylobacteria associated with the hyper accumulating plant Thlaspi goesingense and description of Methylobacterium goesingense sp. nov. Syst Appl Microbiol 29:634–644, DOI 10.1016/j.syapm.2006.01.011
Ivanova EG, Doronina NV, Shepelyakovskaya AO, Laman AG, Brovko FA, Trotsenko YA (2000) Facultative and obligate aerobic methylobacteria synthesize cytokinins. Microbiology 69:646–651, DOI 10.1023/A:1026693805653
Ivanova EG, Doronina NV, Trotsenko YA (2001) Aerobic methylobacteria are capable of synthesizing auxins. Microbiology 70:392–397, DOI 10.1023/A:1010469708107
Jaccard P (1912) The distribution of the flora in the alpine zone. New Phytol 11:37–50, DOI 10.1111/j.1469-8137.1912.tb05611.x
Jourand P, Giraud E, Bena G, Sy A, Willems A, Gillis M et al (2004) Methylobacterium nodulans sp. nov., for a group of aerobic, facultatively methylotrophic, legume root-nodule-forming and nitrogen-fixing bacteria. Int J Syst Evol Microbiol 54:2269–2273, DOI 10.1099/ijs.0.02902-0
Kang YS, Kim J, Shin HD, Nam YD, Bae JW, Jeon CO et al (2007) Methylobacterium platani sp. nov., isolated from a leaf of the tree Platanus orientalis. Int J Syst Evol Microbiol 57:2849–2853, DOI 10.1099/ijs.0.65262-0
Knief C, Frances L, Cantet F, Vorholt JA (2008) Cultivation-independent characterization of methylobacterium populations in the plant phyllosphere by automated ribosomal intergenic spacer analysis. Appl Environ Microbiol 74:2218–2228, DOI 10.1128/AEM.02532-07
Kutschera U (2007) Plant-associated methylobacteria as co-evolved phytosymbionts. Plant Signal Behav 2:74–78
Lambais MR, Crowley DE, Cury JC, Bull RC, Rodrigues RR (2006) Bacterial diversity in tree canopies of the Atlantic forest. Science 312:1917
Lee HS, Madhaiyan M, Kim CW, Choi SJ, Chung KY, Sa TM (2006) Physiological enhancement of early growth of rice seedlings (Oryza sativa L.) by production of phytohormone of N2-fixing methylotrophic isolates. Biol Fertil Soils 42:402–408, DOI 10.1007/s00374-006-0083-8
Lidstrom ME, Chistoserdova L (2002) Plants in the pink: cytokinin production by Methylobacterium. J Bacteriol 184:1818, DOI 10.1128/JB.184.7.1818.2002
Madhaiyan M, Poonguzhali S, Lee HS, Hari K, Sundaram SP, Sa T (2005) Pink-pigmented facultative methylotrophic bacteria accelerate germination, growth and yield of sugarcane clone Co86032 (Saccharum officinarum L.). Biol Fertil Soils 41:350–358, DOI 10.1007/s00374-005-0838-7
Madhaiyan M, Poonguzhali S, Ryu J, Sa T (2006a) Regulation of ethylene levels in canola (Brassica campestris) by 1-aminocyclopropane-1-carboxylate deaminase-containing Methylobacterium fujisawaense. Planta 224:268–278, DOI 10.1007/s00425-005-0211-y
Madhaiyan M, Suresh Reddy BV, Anandam R, Senthilkumar M, Poonguzhali S, Sundaram SP et al (2006b) Plant growth-promoting Methylobacterium induces defense responses in ground nut (Arachis hypogaea L.) compared with root pathogen. Curr Microbiol 53:270–276, DOI 10.1007/s00284-005-0452-9
Madhaiyan M, Kim BY, Poonguzhali S, Kwon SW, Song MH, Ryu JH et al (2007) Methylobacterium oryzae sp. nov., an aerobic, pink-pigmented, facultatively methylotrophic, 1-aminocyclopropane-1-carboxylate deaminase-producing bacterium isolated from rice. Int J Syst Evol Microbiol 57:326–331, DOI 10.1099/ijs.0.64603-0
Margalef R (1958) Information theory in ecology. Gen Syst 3:36–71
McDonald IR, Kenna EM, Murrell JC (1995) Detection of methanotrophic bacteria in environmental samples with PCR. Appl Environ Microbiol 61:116–121
McDonald IR, Doronina NV, Trotsenko YA, McAnulla C, Murrell JC (2001) Hyphomicrobium chloromethanicum sp. nov. and Methylobacterium chloromethanicum sp. nov., chloromethane-utilizing bacteria isolated from a polluted environment. Int J Syst Evol Microbiol 51:119–122
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273, DOI 10.1073/pnas.76.10.5269
Opelt K, Berg C, Schönmann S, Eberl L, Berg G (2007) High specificity but contrasting biodiversity of Sphagnum-associated bacterial and plant communities in bog ecosystems independent of the geographical region. ISME J 1:502–516, DOI 10.1038/ismej.2007.58
Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Patt TE, Cole GC, Hanson RS (1976) Methylobacterium, a new genus of facultatively methylotrophic bacteria. Int J Syst Bacteriol 26:226–229
Pielou EC (1969) An introduction to mathematical ecology. Wiley, London
Raja P, Uma S, Sundaram SP (2006) Non-nodulating pink-pigmented facultative Methylobacterium sp. with a functional nifH gene. World J Microbiol Biotechnol 22:1381–1384, DOI 10.1007/s11274-006-9199-0
Romanovskaya VA, Stolyar SM, Malashenko YR (1996) Occurrence and abundance of bacteria of the genus Methylobacterium in various ecosystems of Ukraine. Mikrobiol Zh 58:3–11
Ryu J, Madhaiyan M, Poonguzhali S, Yim W, Indiragandhi P, Kim K et al (2006) Plant growth substances produced by Methylobacterium spp. and their effect on tomato (Lycopersicon esculentum L.) and red pepper (Capsicum annuum L.) growth. J Microbiol Biotechnol 16:1622–1628
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shannon CE, Weaver W (1949) The mathematical theory of communication. University of Illinois Press, Urbana
Sy A, Giraud E, Jourand P, Garcia N, Willems A, deLajudie P et al (2001) Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J Bacteriol 183:214–220, DOI 10.1128/JB.183.1.214-220.2001
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882, DOI 10.1093/nar/25.24.4876
Trotsenko IA, Ivanova EG, Doronina NV (2001) Aerobic methylotrophic bacteria as phytosymbionts. Microbiol 70:725–736, DOI 10.1023/A:1013167612105
Tsuji K, Tsien HC, Hanson RS, De Palma SR, Scholtz R, LaRoche S (1990) 16S ribosomal RNA sequence analysis for determination of phylogenetic relationship among methylotrophs. J Gen Microbiol 136:1–10
Urakami T, Araki H, Suzuki K, Komogata K (1993) Further studies of the genus Methylobacterium and description of Methylobacterium aminovorans sp. nov. Int J Syst Bacteriol 43:504–513
Van-Aken B, Peres CM, Doty SL, Yoon JM, Schnoor JL (2004) Methylobacterium populi sp. nov., a novel aerobic, pink-pigmented, facultatively methylotrophic, methane-utilizing bacterium isolated from poplar trees (Populus deltoides x nigra DN34). Int J Syst Evol Microbiol 54:1191–1196, DOI 10.1099/ijs.0.02796-0
Wood AP, Kelly DP, McDonald IR, Jordan SL, Morgan TD, Khan S et al (1998) A novel pink-pigmented facultative methylotroph, Methylobacterium thiocyanatum sp. nov., capable of growth on thiocyanate or cyanate as sole nitrogen sources. Arch Microbiol 169:148–158, DOI 10.1007/s002030050554
Yang CH, Crowley DE, Borneman J, Keen NT (2001) Microbial phyllosphere populations are more complex than previously realized. Proc Natl Acad Sci USA 98:3889–3894, DOI 10.1073/pnas.051633898
Acknowledgement
This work was supported by the research project of Indian Council of Agricultural Research, New Delhi through TMC-Mission Mode Project for Sustainable and cost effective production of high quality cotton in fiber. The authors are also thankful to Dr. T. Sa, Chungbuk National University, Republic of Korea and Dr. P. Jourand, Campus International de Baillarguet, France for providing type strains of Methylobacterium species. We are thankful to Lisa Scheper, Florida International University, Maimi, FL, USA for reviewing and valuable suggestions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Raja, P., Balachandar, D. & Sundaram, S.P. Genetic diversity and phylogeny of pink-pigmented facultative methylotrophic bacteria isolated from the phyllosphere of tropical crop plants. Biol Fertil Soils 45, 45–53 (2008). https://doi.org/10.1007/s00374-008-0306-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-008-0306-2