Abstract
Objectives
To automatically detect MRI artifacts on dynamic contrast-enhanced (DCE) maximum intensity projections (MIPs) of the breast using deep learning.
Methods
Women who underwent clinically indicated breast MRI between October 2015 and December 2019 were included in this IRB-approved retrospective study. We employed two convolutional neural network architectures (ResNet and DenseNet) to detect the presence of artifacts on DCE MIPs of the left and right breasts. Networks were trained on images acquired up to and including the year 2018 using a 5-fold cross-validation (CV). Ensemble classifiers were built with the resulting CV models and applied to an independent holdout test dataset, which was formed by images acquired in 2019.
Results
Our study sample contained 2265 examinations from 1794 patients (median age at first acquisition: 50 years [IQR: 17 years]), corresponding to 1827 examinations of 1378 individuals in the training dataset and 438 examinations of 416 individuals in the holdout test dataset with a prevalence of image-level artifacts of 53% (1951/3654 images) and 43% (381/876 images), respectively. On the holdout test dataset, the ResNet and DenseNet ensembles demonstrated an area under the ROC curve of 0.92 and 0.94, respectively.
Conclusion
Neural networks are able to reliably detect artifacts that may impede the diagnostic assessment of MIPs derived from DCE subtraction series in breast MRI. Future studies need to further explore the potential of such neural networks to complement quality assurance and improve the application of DCE MIPs in a clinical setting, such as abbreviated protocols.
Key Points
• Deep learning classifiers are able to reliably detect MRI artifacts in dynamic contrast-enhanced protocol-derived maximum intensity projections of the breast.
• Automated quality assurance of maximum intensity projections of the breast may be of special relevance for abbreviated breast MRI, e.g., in high-throughput settings, such as cancer screening programs.
Similar content being viewed by others
Introduction
Breast cancer is the most common cancer in women. Over the past decades, population-based screening programs have been implemented aiming to detect breast cancer in earlier stages and to reduce mortality rates [1,2,3]. The most widely used diagnostic method in breast cancer screening is X-ray mammography. In contrast to this, magnetic resonance imaging (MRI) has been described in several studies to provide a higher sensitivity with regard to breast cancer detection (e.g., [4,5,6,7]). Using MRI in breast imaging has historically been accompanied by discussions about its potential to contributing to overdiagnosis and even overtreatment [8, 9], as well as with regard to the question whether the increased sensitivity of MRI (with detection of cancer at earlier stages and reduced interval cancer rates) effectively contributes to a survival benefit [10, 11]. The latter, however, is suggested in a recent literature review by Mann et al [12] for participants in the MRI screening studies. These aspects, as well as its high direct and indirect costs, might have contributed to the so far relatively limited widespread use of MRI in, e.g., breast cancer screening, although promising results, for example serving in supplemental screening for women with dense breast tissue, have been reported [11], and also despite beneficial features such as the lack of radiation exposure and the possibility to extract kinetic tissue features such as perfusion [13]. Common clinical indications to perform diagnostic MRI imaging of the breast are therefore limited and include breast cancer screening in high-risk patients and improvement of diagnostic sensitivity in dense breast tissue, among others [14].
In order to further increase the efficiency and feasibility of using MRI for the application in population-based breast cancer screening, the shortening of MRI protocols by reducing both the number of sequences acquired and the assessment time is the focus of ongoing research to develop abbreviated MRI protocols (e.g., [15,16,17,18,19]). Some of these approaches include image visualizations with maximum intensity projections (MIPs) to represent the highest intensity values along one axis of a 3-dimensional (3D) volume in a 2-dimensional (2D) image, allowing radiologists to quickly interpret the whole volume based on this 2D projection. Kuhl et al [15] were among the first to propose an abbreviated MRI protocol for breast cancer screening that included the assessment of dynamic contrast-enhanced (DCE) MIPs derived from subtraction images of postcontrast images with a high negative predictive value (NPV) in their study.
Peculiarities in MRI breast imaging, such as the required large field of view or the anatomy itself, which makes the positioning of the patient in special radiofrequency coils an important task influencing the image quality, make it particularly susceptible to the occurrence of image artifacts [20, 21]. Although many techniques exist to prevent them, artifacts commonly occur in MRI examinations, which can be caused by various reasons [22,23,24,25]. The recognition of artifacts is of high relevance as their presence may significantly impede diagnostic assessment. This is particularly relevant when applying DCE protocols, since contrast agent application cannot be repeated during a single examination, thus needing for an additive repeat examination and a double exposure to contrast agents in case of low image quality.
Therefore, additional methods for assessing artifacts would be desirable to inform radiologists about their presence when evaluating MIPs, e.g., in the context of abbreviated breast imaging protocols where limited sequences are acquired and artifacts might not be compensated by complementary sequences. We propose a deep learning–based approach for an automated detection of MRI artifacts on DCE MIPs to support and improve the diagnostic assessment. Therefore, we trained two convolutional neural network (CNN) algorithms to binary classify MIP images of the left and right breasts with regard to the presence of significant artifacts.
Material and methods
Study sample and ethics approval
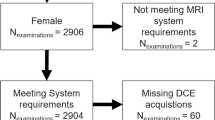
The retrospective study was approved by the ethics committee of the Friedrich-Alexander-University Erlangen-Nürnberg, waiving the need for informed consent. Women who underwent clinically indicated breast MRI at the Institute of Radiology of the University Hospital Erlangen (UHE) between October 2015 and December 2019 were included in this study. Inclusion was performed independent of the respective indications, which covered all current clinical indications for a breast MRI examination. Only entirely completed MRI examinations acquired with a full diagnostic protocol including contrast agent application were included in the analysis.
MRI protocol
The MRI examinations were performed using the routine clinical MRI devices (1.5–3 Tesla MRI; model names: Aera, Avanto, Sola, Vida, and Skyra from Siemens Healthineers). The clinical MRI protocol consisted of morphologic, contrast-enhanced, and diffusion-weighted imaging (DWI) sequences. Herein, morphologic, non-contrast-enhanced MRI sequences included T2-weighted sequences with and without fat suppression, dynamically acquired T1-weighted imaging sequences, and DWI sequences. Subtraction series were created by subtracting the second postcontrast images from the native T1-weighted images by the scanner system. A detailed overview of the range of different DCE sequence settings is given in supplemental Table S1.
Data processing
All data were transferred from the routine picture archiving and communication system to analytic workstations. For each included subtraction volume, a MIP of the DCE subtraction image was generated in direction of the slice axis, which was identical to the z-axis in all datasets, resulting in a 2D representation of voxels with the highest intensities along the transverse plane. From these MIPs, the upper left and right image quadrants were cropped out automatically using Python (version 3.8.5), displaying the left and right breasts as regions of interest (ROIs). The cropping was assisted by anatomical structures based on the sternum position, which was derived from the corresponding native T1-weighted series.
Visual artifact assessment
One radiologist (S.B. > 10 years of experience in radiology) visually rated the processed images with regard to the presence of artifacts. A binary labeling was performed on the level of individual MIP images of the left and right breasts, indicating either the presence of (one or more) artifacts (1 = artifacts present) or the absence of any artifacts (0 = no artifacts present), without further localizing the artifacts on the images. The positive class was defined as an artifact with the potential to mask a suspicious finding of any size on the image but did not have to occur within the area of an existing suspicious lesion in MRI examinations that presented with a suspicious lesion visible on the MIP.
Image preprocessing and image augmentation
All preprocessing steps were performed in Python (version 3.8.5) using the SimpleITK library version 2.0.2 [26, 27]. A MIP was computed from each included subtraction volume of the second postcontrast phase in the direction of the z-axis resulting in a 2D representation of voxels with the highest intensities along the transverse plane. We here used images of the second postcontrast phase as related to the timing of the contrast agent administration in the locally established MRI protocols; this postcontrast phase is considered to provide the highest image quality. From the resulting images, the upper left and right image quadrants were cropped out, displaying the left and right breasts as ROIs (see schema in the supplementary information, Figure S1). The cropping was assisted by anatomical structures based on the sternum position, which was derived from the corresponding native T1-weighted series using an in-house developed Python script. This sternum-assisted cropping was applied only if the cropping position ycrop was greater than 1/2 × imageheight to ensure the inclusion of the ROI in the resulting image section. The exact cropping position ycrop was calculated by adding an offset of 5% of the image height to the sternum position:
Images were further normalized (mean = 0, standard deviation = 1), resized to 256 × 256 pixel, and saved as NumPy arrays [28] that served as input for the deep learning algorithms. For the visual expert reading regarding the presence of artifacts, these cropped images were additionally saved as JPEG files. The following standard image augmentation techniques were implemented using the monai Python library version 0.4.0 [29]: random rotation (probability: 0.5; maximum angle: 180 degrees), random flip across x-axis and y-axis (probability: 0.5), and random zoom (probability: 0.5; minimum zoom: 0.5; maximum zoom: 1.5).
Deep learning
Experiments were performed using two CNN architectures, a DenseNet121 [30] and a ResNet18 [31], to perform a binary classification to detect the presence or absence of artifacts on DCE MIP images of the left and right breasts. The data were split into a training dataset formed by the examinations acquired up to and including the year 2018 and an independent holdout test dataset including all examinations acquired in 2019 from patients, which were not already contained in the training dataset. The neural networks were trained using a stratified 5-fold cross-validation (CV), i.e. a proportion of 20% of the training dataset was used for testing in each fold. The training data of each fold was further split by 80 to 20% into the actual training dataset and a validation dataset to monitor the validation loss and metrics. All CV models were trained for 200 epochs using a batch size of 128, resulting in 19 steps per epoch. Trainings were carried out on a Tesla V100 graphics processing unit with 32GB memory and an Intel® Xeon® CPU E5-2698 v4 @2.20GHz (20 cores) with 256GB RAM.
For each network architecture, an ensemble classifier was built from the CV models using the weights from the epoch with the lowest validation loss observed within 200 epochs. Hence, each of the two ensembles consisted of 5 models, which were finally applied to predict the presence of artifacts in the holdout test dataset ; the predicted probabilities of the 5 models were averaged [32] and images with final probabilities > 0.5 were considered to contain artifacts. Further details on the initial network settings and network modifications are given in the supplementary information (the “Deep Learning” section).
Statistical analysis
Statistical analyses were performed with the R software version 4.0.4 [33]. Continuous variables were compared between two groups using the Wilcoxon rank sum test (two-sided). Relationships between categorical variables were assessed by measuring their association in contingency tables. The calculation of basic summary statistics and the Wilcoxon rank sum test [34] was implemented in base R [33]. Contingency tables (with statistics) were calculated with the sjstats R package version 0.18.1 [35]. The φ correlation coefficient [36] was used to measure the association between two dichotomous variables. Its p value was calculated using the chi-square test. For all statistical tests, a significance level α = 0.05 was used. Model metrics were calculated with the mlr3measures package version 0.3.1 [37]. Graphics were created with the R packages ggplot2 version 3.3.3 [38], ggpubr version 0.4.0 [39], and precrec version 0.12.1 [40]. To assess the strength of agreement between the ResNet and the DenseNet ensemble classifiers with regard to the prediction of artifacts in the independent holdout test dataset, Cohen’s kappa [41] for two raters was calculated using the R package irr version 0.84.1 [42]. According to Landis and Koch, the strength of agreement based on the kappa statistic can be categorized as follows: kappa < 0.00, poor; 0.00 to 0.20, slight; 0.21 to 0.40, fair; 0.41 to 0.60, moderate; 0.61 to 0.80, substantial; 0.81 to 1.00, almost perfect [43]. To support the interpretability of the deep learning classifiers, class activation maps (CAMs) can be used to represent so-called discriminative regions as color-coded heatmaps, which mark locations in the image that are considered important by the CNN classifier to decide on the derived class [44]. These CAMs were computed for the test images from the model with the highest area under the receiver operating characteristic (ROC) curve on the holdout test dataset using the GradCAM++ algorithm [45] to provide a better interpretability of the model’s predictions utilizing the implementation already provided with the monai library [29].
Results
Study sample characteristics
Our study sample contained 2265 MRI examinations from 1794 patients (median age at first acquisition: 50 years [IQR: 17 years]), which were acquired between October 2015 and December 2019. One thousand four hundred sixty-one individuals of the study sample received one MRI examination, 225 individuals received two, 80 individuals received three, and 28 individuals received four or more MRI examinations. The training dataset included examinations acquired up to and including the year 2018, corresponding to 1827 examinations of 1378 patients (median age at first acquisition: 50 years [IQR: 16.75 years]), resulting in a total of 3654 training images. The independent holdout test dataset was formed with all examinations acquired in 2019 from patients, which were not already included in the training dataset. This holdout test dataset contained 438 examinations of 416 patients (median age at first acquisition: 51 years [IQR: 18 years]), resulting in 876 test images. Demographic data, sample characteristics, and target class distribution across the datasets are shown in Table 1.
No significant difference in the distribution of the age at first acquisition could be observed between the training cohort and the test cohort (p value: 0.2). When including repeated studies for one patient, the overall training cohort was significantly younger than the test cohort (p value: < 0.001).
Presence of artifacts within the dataset
Artifacts were detected by the visual reading in 51% (2332 out of 4530 images) of all images in the dataset. This corresponds to the presence of artifacts bilaterally in 36.7% (n = 832 examinations), unilaterally in 29.5% (n = 668 examinations), and the absence of artifacts in 33.8% (n = 765 examinations) of all examinations.
In the training dataset and in the test datasets, artifacts were present in 53% (1951 out of 3654 images) and 43% (381 out of 876 images) of all images, respectively, which corresponds to a statistically significant difference of the presence of artifacts between the training dataset and the test dataset (φ correlation coefficient: 0.078; p value: < 0.001).
Deep learning
The ResNet ensemble demonstrated an area under the ROC curve of 0.923 for the detection of artifacts in breast DCE MIPs, while the DenseNet ensemble provided an area under the ROC curve of 0.940 on the holdout test dataset (Table 2). Herein, the NPV was 0.874 for the ResNet and 0.915 for the DenseNet ensemble with nearly equivalent positive predictive values (PPVs) of 0.816 and 0.8, respectively (Table 2).
During the 5-fold CV training, the ResNet models provided an area under the ROC curve of 0.879 (± 0.010) on average and the DenseNet models provided an area under the ROC curve of 0.896 (± 0.012) with an average NPV of 0.776 (± 0.012) and 0.791 (± 0.019), and a PPV of 0.823 (± 0.018) and 0.830 (± 0.029), respectively (Table 3). With a kappa = 0.83 (p < 0.001), there was an “almost perfect” agreement (according to [43]) between the DenseNet and the ResNet ensemble classifiers with regard to the prediction of artifacts in the independent holdout test dataset. Table 4 shows the results of the comparison of the model performance between the DenseNet and the ResNet during CV. For all performance measures, no statistically significant differences between the two network architectures could be observed. However, the number of epochs until convergence (“Best epoch”) and the average time per epoch until convergence were significantly lower/shorter for ResNet compared to DenseNet.
Figure 1 shows the ROC curve (left column) and the precision-recall (PR) curve (mid column) of the 5 ResNet CV models (row 1) and the performance of the ResNet ensemble on the full holdout test dataset (row 2). The training and validation loss curves for the ResNet averaged over the 5 CV folds are shown in the right column of Fig. 1. It can be recognized here that there is no further improvement in the validation loss from about epoch 100, which is in accordance with the best epochs shown in Table 3 (mean of the best epochs of the 5 ResNet CV folds: 88.800 (± 25.371)) and might be caused by overfitting.
ResNet plots. The figure shows the receiver operating characteristic (ROC) curve (left column) and the precision-recall curve (mid column) and the loss curves (right column) for the ResNet architecture. Row 1: ROC and PR curve averaged over 5 cross-validation folds. Row 2: ROC and PR curve for the ensemble’s prediction on the independent holdout test dataset. The training loss (dark blue) and the validation loss (yellow) curves are averaged over 5 CV folds
Figure 2 shows the ROC curve (left column) and the PR curve (mid column) of the 5 DenseNet CV models (row 1) and the performance of the DenseNet ensemble on the full holdout test dataset (row 2). The training and validation loss curves for the DenseNet averaged over the 5 CV folds are shown in the right column of Fig. 2. Here, overfitting can be observed from around epoch 160, which is also reflected by the best epochs shown in Table 3 (mean of the best epochs of the 5 DenseNet CV folds: 159.0 (± 36.407)), however, accompanied by a larger standard deviation compared to the ResNet CV models.
DenseNet plots. The figure shows the receiver operating characteristic (ROC) curve (left column) and the precision-recall curve (mid column) and the loss curves (right column) for the DenseNet architecture. Row 1: ROC and PR curve averaged over 5 cross-validation folds. Row 2: ROC and PR curve for the ensemble’s prediction on the independent holdout test dataset. The training loss (dark blue) and the validation loss (yellow) curves are averaged over 5 CV folds
Class activation maps
Figure 3 exemplarily demonstrates CAMs for one each of a true positive, true negative, false positive, and false negative predicted images from the holdout test dataset for the respective class predicted by the classifier (supplemental Figures S2 to S5 provide further CAM images for each of these categories). All CAM images were computed using the DenseNet model with the highest area under the ROC curve on the holdout test dataset, i.e. CV model M2 (AUROC = 0.938; see supplemental Table S2) and examined and interpreted by an experienced radiologist (S.B.). The class-discriminative regions for the correctly predicted artifact-containing images coincide well with the artifact-affected regions (true positives, Fig. 3 [TP] and supplemental Figure S2). For the correctly predicted artifact-free images, the class-discriminative regions for the negative class rather seem to reflect areas with a sharp demarcation of contrast agent-containing blood vessels from breast tissue (true negatives, Fig. 3 [TN] and supplemental Figure S3). When incorrectly predicting an artifact in artifact-free images (false positives), the CAMs of the (falsely predicted) positive class seem to correlate with either regions in the image that give a blurred impression (Fig. 3 [FP] and supplemental Figure S4, images A and E), or with regions that contain high intensity values (supplemental Figure S4, images B, C, and D), whereas the incorrect classification of the absence of an artifact in an artifact-containing image (false negatives) results in heatmaps for the (falsely predicted) negative class that seems to correlate with regions in the image that contain either high intensity values, such as contrast enhancements spots, or dense breast tissue (Fig. 3 [FN] and supplemental Figure S5, images A and B), and areas with a sharp demarcation of contrast agent-containing blood vessels from breast tissue (supplemental Figure S5, images C–E).
Class activation maps (examples). The figure shows the Grad-CAM++ visualizations for one each of a true positive (TP), true negative (TN), false positive (FP), and false negative (FN) predicted images from the holdout test dataset for the respective predicted class. The heatmaps depict with the color gradient from blue to red the relevance of each pixel for the inference of the respective class.
Artifacts in images with significant lesions
Figure 4 represents 9 clinical cases taken from our dataset with BI-RADS 5 and BI-RADS 6 lesions, respectively. The first row represents 3 different cases without artifacts (a–c). Tiles d–f show images, where the artifact has no or a moderate influence on the diagnostic assessment, whereas in the last row, the presence of artifacts significantly impedes the diagnostic evaluation of the lesions (g–i).
BI-RADS 5 and BI-RADS 6 lesions in clinical cases (examples). Each tile of the figure presents the left or right breast of one clinical case with a diagnosed BI-RADS 5 or BI-RADS 6 lesion. Row 1 (a–c) shows images without the presence of artifacts (a: BI-RADS 6; b: BI-RADS 6; c: BI-RADS 5). Row 2 (d–f) shows images that contain artifacts with no or moderate influence on the diagnostic assessment (d: BI-RADS 5; d: BI-RADS 5; f: BI-RADS 6). Row 3 (g–i) shows images with artifacts potentially impeding the diagnostic evaluation (g: BI-RADS 6; h: BI-RADS 6; i: BI-RADS 5)
Discussion
We demonstrated a deep learning–based approach consisting of two CNN ensembles, each trained with a 5-fold CV, to automatically detect MRI artifacts on DCE MIPs of the breast. The DenseNet ensemble (area under ROC: 0.940) outperformed the ResNet ensemble (area under ROC: 0.923) on the independent holdout dataset (Table 2). While the PPVs derived on the holdout test dataset of both ensemble networks are quite similar (both ~0.8), the NPV of the DenseNet is 0.915 compared to 0.874 for the ResNet. These values suggest that the DenseNet detected artifacts in the independent holdout test dataset quite reliably.
The reasons for artifacts in MRI examinations vary widely with numerous possible sources [22,23,24,25]. Breast MRI itself provides a certain range of potential artifacts that has extensively been examined in literature, also giving advice on possible counter mechanisms [20]. Our results demonstrate that visually detectable artifacts frequently occur in dynamic breast MRI with about two-third of all examinations of our cohort revealing artifacts in DCE MIPs. Artifact prevalence has mainly been investigated to the specific subgroup of motion artifacts, which have been reported by Carbonaro et al [46] with a rate of 35%, whereas Clauser et al [47] reported motion artifacts alone in about 46% of the cases, and Fiaschetti et al [48] reported any artifacts in 33% of the evaluated studies. However, since the exact definition of “artifacts” differs among the studies as do the used MRI devices, the comparability of these results to ours may be limited.
Abbreviated breast MRI protocols with the assessment of MIPs as the primary analysis have been introduced by Kuhl et al [15] in their landmark paper in 2014. With the potential to reduce examination and reading times, the ongoing work to develop abbreviated imaging protocols aims to further increase the applicability of MRI as a highly sensitive method, e.g., for breast cancer screening programs (reviewed in [49]). Besides a majority of MRI breast imaging protocols that include DCE sequences, also contrast-agent free techniques such as DWI are being explored [50]. All abbreviated imaging techniques do share the common feature that only few acquisitions are available for the assessment, further increasing the importance of persistent high image quality since no compensating complementary additive acquisitions are available. Using MIPs as a primary reading source adds up to the challenge, as hyperintense artifacts in the single slices progress into the MIP projections. Thus, for radiologists, the awareness of the presence of artifacts is of high relevance as they can obscure relevant lesions (e.g., Fig. 4, tiles g–i).
For a better interpretability of the neural networks’ decision, we decided to generate CAMs. These represent class-discriminative regions as heatmaps for an input image to visualize image regions that were considered important by the neural network to infer the predicted class [44]. Notably, the class-discriminative regions for the correctly classified artifact images coincide well with image regions that indeed contain artifacts (Fig. 3 [TP] and supplemental Figure S2). CAMs of the correctly classified artifact-free images indicate that a sharp demarcation of contrast agent-containing blood vessels from breast tissue seems to be considered important by the neural network for its decision regarding the absence of artifacts (Fig. 3 [TN] and supplemental Figure S3), which is underlined by the observation that these sharp demarcation of blood vessels may also be related to the occurrence of false negative predictions (Fig. 3 [FN] and supplemental Figure S5). This is reasonable, since this clear differentiation would no longer be given in case of, e.g., the presence of motion artifacts. However, image regions that give a blurred impression or regions with high intensity values may cause false positive predictions (Fig. 3 [FP] and supplemental S4). It has to be noted here that some of the images of supplemental Figure S4 indeed contain slightly blurred image impressions in the areas that are located by the class-discriminative regions in the CAMs for the “falsely predicted” class (rows 2–3 in supplemental Figure S4, images A, C, D, E), which, however, were not considered “significant artifacts” (with the potential to mask a suspicious finding) during the radiologist’s reading.
Recently, researchers also applied artificial intelligence to detect and classify lesions in DCE MIPs of the breast [51,52,53]. Possibly, such automated evaluations might also be hampered by MRI artifacts present on MIP images. Therefore, our results may also help to further improve automated lesion detection and classification by complementing these with an automated artifact detection in the future.
Limitations
Our study has several limitations. First, abbreviated breast MRI protocols are currently mostly evaluated as potential screening examinations; however, our dataset was extracted from a patient population of a university hospital; thus, screening collectives might present different patient characteristics. Second, a significant deviation of the proportion of artifacts was observed between the training and the test dataset (53% vs. 43% artifact-containing images, respectively). We are aware of the fact that the neural networks were exposed to a different class distribution during training compared to inferring the holdout test dataset, potentially influencing their performance. Third, another limitation could be the use of a binary outcome in our present study, potentially leading to images classified as “negative,” which may, however, contain slight artifacts considered insignificant to the image evaluation (as outlined above). In contrast, artifact-containing images were not further subcategorized, resulting in some range of artifact severity and different types of artifacts in images of the positive class. Since these circumstances could contribute to the occurrence of both false positives and false negatives, future studies could include a more finely granulated artifact categorization. In addition, since the analysis was based on artifacts in MIP images, artifacts or technical issues that might impede the diagnostic assessment but are invisible on MIPs might have been missed, e.g., an improper administration of contrast agents. Another important limitation is that the information on the scanner model and magnetic field strength were not provided as input for training the deep learning algorithms. A subsequent analysis of the ensemble classifiers’ model performance on the holdout test dataset stratified by scanner model (supplementary information, Tables S3 and S4) indicates that these information might indeed be helpful for improving the predictions and future studies should include them to investigate their influence on the prediction of artifacts in MRI images. There is also some evidence that the presence of lesions (defined as BI-RADS score ≥ 3) may potentially influence the classifiers’ performance (supplementary information, Tables S5 and S6). It would be interesting to evaluate this in more detail in the future, although the practical use in artifact detection for quality assurance is certainly limited, as the information on the BI-RADS score would need to be available in advance of the diagnostic MRI examination. Furthermore, although we have tested the algorithms on an unseen and independent holdout test dataset, no external validation was performed in this retrospective single institution study. Finally, a multi-reader setting to establish the ground truth might further improve the automated artifact detection in MIPs.
Conclusion
In summary, neural networks are able to reliably detect artifacts that may impede the diagnostic assessment of MIPs derived from DCE subtractions series in breast MRI protocols. Although future studies are required to further improve the detection of artifacts in MRI images using deep learning and to investigate the relevance of these methods to complement quality assurance in the clinical settings, our work demonstrates the potential of neural networks to serve as quality indicators and to complement automated lesion detection and classification.
Abbreviations
- AUPRC:
-
Area under the precision-recall curve
- AUROC:
-
Area under the receiver operating characteristic curve
- CAM:
-
Class activation map
- CNN:
-
Convolutional neural network
- CPU:
-
Central processing unit
- CV:
-
Cross-validation
- DCE:
-
Dynamic contrast enhanced
- DWI:
-
Diffusion-weighted imaging
- GB:
-
Gigabyte
- IQR:
-
Interquartile range
- IRB:
-
Institutional review board
- MIP:
-
Maximum intensity projection
- MRI:
-
Magnetic resonance imaging
- NPV:
-
Negative predictive value
- PPV:
-
Positive predictive value
- PR:
-
Precision-recall
- RAM:
-
Random-access memory
- ROC:
-
Receiver operating characteristic
- ROI:
-
Region of interest
- T:
-
Tesla
- UHE:
-
University Hospital Erlangen
- X-ray:
-
Röntgen radiation
References
Veronesi U, Boyle P, Goldhirsch A, Orecchia R, Viale G (2005) Breast cancer. Lancet 365:1727–1741
Oeffinger KC, Fontham ET, Etzioni R et al (2015) Breast cancer screening for women at average risk: 2015 guideline update from the American Cancer Society. JAMA 314:1599–1614
Untch M, Fasching PA, Brucker SY et al (2021) Treatment of patients with early breast cancer: evidence, controversies, consensus: German Expert Opinions on the 17th International St. Gallen Consensus Conference. Geburtshilfe Frauenheilkd 81:637–653
Kriege M, Brekelmans CTM, Zonderland HM, Kok T, Meijer S (2004) Efficacy of MRI and mammography for breast-cancer screening in women with a familial or genetic predisposition. N Engl J Med 351:427–437
Kuhl CK, Schrading S, Leutner CC et al (2005) Mammography, breast ultrasound, and magnetic resonance imaging for surveillance of women at high familial risk for breast cancer. J Clin Oncol 23:8469–8476
Peters NH, Borel Rinkes IH, Zuithoff NP, Mali WP, Moons KG, Peeters PH (2008) Meta-analysis of MR imaging in the diagnosis of breast lesions. Radiology 246:116–124
Weinstein SP, Localio AR, Conant EF, Rosen M, Thomas KM, Schnall MD (2009) Multimodality screening of high-risk women: a prospective cohort study. J Clin Oncol 27:6124–6128
Evans A, Vinnicombe S (2017) Overdiagnosis in breast imaging. Breast 31:270–273
Jatoi I, Pinsky PF (2021) Breast cancer screening trials: endpoints and overdiagnosis. J Natl Cancer Inst 113:1131–1135
Morrow M, Waters J, Morris E (2011) MRI for breast cancer screening, diagnosis, and treatment. Lancet 378:1804–1811
Bakker MF, de Lange SV, Pijnappel RM et al (2019) Supplemental MRI screening for women with extremely dense breast tissue. N Engl J Med 381:2091–2102
Mann RM, Kuhl CK, Moy L (2019) Contrast-enhanced MRI for breast cancer screening: breast MRI for Screening. J Magn Reson Imaging 50:377–390
Jaglan P, Dass R, Duhan M (2019) Breast cancer detection techniques: issues and challenges. J Inst Eng India Ser B 100:379–386. https://doi.org/10.1007/s40031-019-00391-2
Krassuski LM, Kautz-Freimuth S, Vennedey V, Rhiem K, Schmutzler RK, Stock S (2021) Decision aids for preventive treatment alternatives for BRCA1/2 mutation carriers: a systematic review. Geburtshilfe Frauenheilkd 81:679–698
Kuhl CK, Schrading S, Strobel K, Schild HH, Hilgers R-D, Bieling HB (2014) Abbreviated breast magnetic resonance imaging (MRI): first postcontrast subtracted images and maximum-intensity projection - a novel approach to breast cancer screening with MRI. J Clin Oncol 32:2304–2310
Grimm LJ, Soo MS, Yoon S, Kim C, Ghate SV, Johnson KS (2015) Abbreviated screening protocol for breast MRI. Acad Radiol 22:1157–1162
Chen S-Q, Huang M, Shen Y-Y, Liu C-L, Xu C-X (2017) Abbreviated MRI protocols for detecting breast cancer in women with dense breasts. Korean J Radiol 18:470–475
Chhor CM, Mercado CL (2017) Abbreviated MRI protocols: wave of the future for breast cancer screening. AJR Am J Roentgenol 208:284–289
Deike-Hofmann K, Koenig F, Paech D et al (2019) Abbreviated MRI protocols in breast cancer diagnostics: abbreviated breast MRI. J Magn Reson Imaging 49:647–658
Harvey JA, Hendrick RE, Coll JM, Nicholson BT, Burkholder BT, Cohen MA (2007) Breast MR imaging artifacts: how to recognize and fix them. Radiographics 27:131–145
Yitta S, Joe BN, Wisner DJ, Price ER, Hylton NM (2013) Recognizing artifacts and optimizing breast MRI at 1.5 and 3 T. AJR Am J Roentgenol. https://doi.org/10.2214/AJR.12.10013
Zhuo J, Gullapalli RP (2006) MR artifacts, safety, and quality control. Radiographics 26:275–297
Smith TB, Nayak KS (2010) MRI artifacts and correction strategies. Imaging Med 2:445–457
Krupa K, Bekiesinska-Figatowska M (2015) Artifacts in magnetic resonance imaging. Pol J Radiol 80:93–106
Budrys T, Veikutis V, Lukosevicius S, Gleizniene R, Monastyreckiene E, Kulakiene I (2018) Artifacts in magnetic resonance imaging: how it can really affect diagnostic image quality and confuse clinical diagnosis? Journal of Vibroengineering 20:1202–1213
Lowekamp BC, Chen DT, Ibanez L, Blezek D (2013) The design of SimpleITK. Front. Neuroinform. https://doi.org/10.3389/fninf.2013.00045
Yaniv Z, Lowekamp BC, Johnson HJ, Beare R (2018) SimpleITK image-analysis notebooks: a collaborative environment for education and reproducible research. J Digit Imaging 31:290–303
Harris CR, Millman KJ, van der Walt SJ et al (2020) Array programming with NumPy. Nature 585:357–362. https://doi.org/10.1038/s41586-020-2649-2
T.M. Consortium (2020) Project MONAI. https://doi.org/10.5281/zenodo.4323059
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks, in: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE Honolulu, HI. https://doi.org/10.1109/CVPR.2017.243
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition, in: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE Las Vegas, NV, USA. https://doi.org/10.1109/CVPR.2016.90
J. Kittler, and F. Roli, eds. (2000) Multiple classifier systems. Springer Berlin
R Core Team (2021) R: a language and environment for statistical computing R Foundation for Statistical Computing Vienna, Austria. https://www.R-project.org/
Bauer DF (1972) Constructing confidence sets using rank statistics. Journal of the American Statistical Association 67:687–690
D. Lüdecke (2021) Sjstats: statistical functions for regression models (version 0.18.1). https://doi.org/10.5281/zenodo.1284472
Warrens MJ (2008) On association coefficients for 2x2 tables and properties that do not depend on the marginal distributions. Psychometrika 73:777–789. https://doi.org/10.1007/s11336-008-9070-3
M. Lang (2021) Mlr3measures: performance measures for ’Mlr3’. https://CRAN.R-project.org/package=mlr3measures
H. Wickham (2016) Ggplot2: elegant graphics for data analysis. http://ggplot2.tidyverse.org
A. Kassambara (2020) Ggpubr: ’ggplot2’ based publication ready plots. https://CRAN.R-project.org/package=ggpubr
Saito T, Rehmsmeier M (2017) Precrec: fast and accurate precision-recall and ROC curve calculations in R. Bioinformatics 33:145–147
Cohen J (1960) A coefficient of agreement for nominal scales. Educational and Psychological Measurement 20:37–46
M. Gamer, J. Lemon, and I.F.P. Singh (2019) Irr: various coefficients of interrater reliability and agreement. https://CRAN.R-project.org/package=irr
Landis JR, Koch GG (1977) The measurement of observer agreement for categorical data. Biometrics 33:159 https://www.jstor.org/stable/2529310
Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A (2016) Learning deep features for discriminative localization, in: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE Las Vegas, NV, USA. https://doi.org/10.1109/CVPR.2016.319
Chattopadhay A, Sarkar A, Howlader P, Balasubramanian VN (2018) Grad-CAM++: generalized gradient-based visual explanations for deep convolutional networks, in: 2018 IEEE Winter Conference on Applications of Computer Vision (WACV). IEEE Lake Tahoe, NV. https://doi.org/10.1109/WACV.2018.00097
Carbonaro LA, Schiaffino S, Clauser P et al (2021) Side of contrast injection and breast size correlate with motion artifacts grade and image quality on breast MRI. Acta Radiol 62:19–26
Clauser P, Dietzel M, Weber M, Kaiser CG, Baltzer PA (2019) Motion artifacts, lesion type, and parenchymal enhancement in breast MRI: what does really influence diagnostic accuracy? Acta Radiol 60:19–27
Fiaschetti V, Pistolese C, Funel V et al (2013) Breast MRI artefacts: evaluation and solutions in 630 consecutive patients. Clin Radiol 68:601–608
Leithner D, Moy L, Morris EA, Marino MA, Helbich TH, Pinker K (2019) Abbreviated MRI of the breast: does it provide value? J Magn Reson Imaging 49:e85–e100. https://doi.org/10.1002/jmri.26291
Bickelhaupt S, Laun FB, Tesdorff J et al (2016) Fast and noninvasive characterization of suspicious lesions detected at breast cancer X-ray screening: capability of diffusion-weighted MR Imaging with MIPs. Radiology 278:689–697
Antropova N, Abe H, Giger ML (2018) Use of clinical MRI maximum intensity projections for improved breast lesion classification with deep convolutional neural networks. J Med Imaging (Bellingham) 5:0145031–0145036. https://doi.org/10.1117/1.JMI.5.1.014503
Adachi M, Fujioka T, Mori M et al (2020) Detection and diagnosis of breast cancer using artificial intelligence based assessment of maximum intensity projection dynamic contrast-enhanced magnetic resonance images. Diagnostics (Basel) 10:330
Hu Q, Whitney HM, Li H, Yu J, Liu P, Giger ML (2021) Improved classification of benign and malignant breast lesions using deep feature maximum intensity projection MRI in breast cancer diagnosis using dynamic contrast-enhanced MRI. Radiol Artif Intell. https://doi.org/10.1148/ryai.2021200159
Acknowledgements
We thank the d.hip Digital Health Innovation Platform for their support. We thank Martin Gründl (Medical Center for Information and Communication Technology, Universitätsklinikum Erlangen) for his support in providing the data, and the Data Integration Center (Medical Center for Information and Communication Technology, Universitätsklinikum Erlangen) for their support.
Funding
Open Access funding enabled and organized by Projekt DEAL. The authors state that this work has received funding (in part) from the German Federal Ministry of Education and Research (BMBF) in association with the project SMART SELECT MRI under the funding number: FKZ 161B0976.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
Institutional review board approval was obtained.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Conflict of interest
As the clinical routine MRI systems in the hospital are manufactured by the company Siemens Healthcare GmbH, the following individual COIs are disclosed. The University Hospital Erlangen receives funding from Siemens Healthcare GmbH that is partially contributing to the job position of Lorenz Kapsner. Sebastian Bickelhaupt received lecture fees from Siemens Healthcare GmbH, and holds (pending) patent applications in MRI; the University Hospital Erlangen receives funding from Siemens Healthcare GmbH that is partially contributing to the job position. Michael Uder is part of the Speakers Bureau of the Siemens Healthcare GmbH.
Guarantor
The scientific guarantor of this publication is PD Dr. med. Sebastian Bickelhaupt.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Methodology
• retrospective
• observational
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kapsner, L.A., Ohlmeyer, S., Folle, L. et al. Automated artifact detection in abbreviated dynamic contrast-enhanced (DCE) MRI-derived maximum intensity projections (MIPs) of the breast. Eur Radiol 32, 5997–6007 (2022). https://doi.org/10.1007/s00330-022-08626-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-08626-5