Abstract
Objective
To compare the robustness of native T1 mapping using mean and median pixel-wise quantification methods.
Methods
Fifty-seven consecutive patients without overt signs of heart failure were examined in clinical routine for suspicion of cardiomyopathy. MRI included the acquisition of native T1 maps by a motion-corrected modified Look-Locker inversion recovery sequence at 1.5 T. Heart function status according to four established volumetric left ventricular (LV) cardio MRI parameter thresholds was used for retrospective separation into subgroups of normal (n = 26) or abnormal heart function (n = 31). Statistical normality of pixel-wise T1 was tested on each myocardial segment and mean and median segmental T1 values were assessed.
Results
Segments with normally distributed pixel-wise T1 (57/58%) showed no difference between mean and median quantification in either patient group, while differences were highly significant (p < 0.001) for the respective 43/42% non-normally distributed segments. Heart function differentiation between two patient groups was significant in 14 myocardial segments (p < 0.001–0.040) by median quantification compared with six (p < 0.001–0.042) by using the mean. The differences by median quantification were observed between the native T1 values of the three coronary artery territories of normal heart function patients (p = 0.023) and insignificantly in the abnormal patients (p = 0.053).
Conclusion
Median quantification increases the robustness of myocardial native T1 definition, regardless of statistical normality of the data. Compared with the currently prevailing method of mean quantification, differentiation between LV segments and coronary artery territories is better and allows for earlier detection of heart function impairment.
Key Points
• Median pixel-wise quantification of native T1 maps is robust and can be applied regardless of the statistical distribution of data points.
• Median quantification is more sensitive to early heart function abnormality compared with mean quantification.
• The new method yields significant native T1 value differentiation between the three coronary artery territories.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pre-contrast T1 relaxation time, the parameter at stake in native T1 mapping, has shown its potential for identifying myocardial tissue abnormality [1], with the limitation that the values measured are sequence-specific [2,3,4,5,6,7]. Native T1 increases may indicate disease and have been associated with diffuse myocardial fibrosis in different types of cardiomyopathy [7,8,9,10,11,12,13,14,15]. Moreover, in patient groups with myocardial impairment, an increase of native T1 was observed in the absence of late gadolinium enhancement (LGE) [7, 9, 10, 14, 15] suggesting that native T1 mapping can be an early indicator of myocardial tissue abnormality. Therefore, a robust native T1 quantification method is needed to ensure early identification of heart function abnormality.
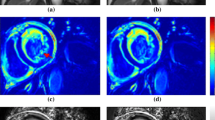
In measuring cardiac T1 value, numerous studies showed normal native T1 variation on different myocardial regions [4, 5, 8, 10, 11, 13, 16,17,18,19]. Intersegmental variations complicate the standardisation of normal values and disease identification. Pixel-wise T1 value quantification also faces variability due to protocol parameters, sequence design, scanner adjustment, T1 fit model, tissue characteristics, and patient’s condition [6, 20]. In view of the heterogeneity of pixel-wise T1 values as illustrated in Fig. 1, variability may be reduced by the assessment of median values of pixel-wise T1 per segment rather than the evaluation of the means [14].
In liver and heart iron deposition assessment by T2* mapping, pixel-wise median quantification produced lower observer variability compared with mean quantification [21] and lower T2* variability in different myocardial regions [22, 23]. These studies showed that partial volume effect, heart motion artefact, the fitting model used, and observer’s myocardial contour determination influence the pixel-wise assessment and quantification in the region of interest. However, pixel-wise native T1 assessment studies published to date used mean quantification with a few ones checking the normality of the statistical distribution of datasets as a whole [11, 13,14,15, 24] rather than performing statistical normality testing of pixel-wise T1 distribution per segment. This study aims to investigate the normality of pixel-wise T1 values per left ventricular heart segment and subsequently compare the mean and median values. Application of both methods on patients with normal and abnormal heart function is used to assess their potential for early detection of heart function abnormality.
Materials and methods
This retrospective analysis was conducted on magnetic resonance imaging (MRI) data acquired from May until October 2015 with approval by the hospital review board waiving the requirement of informed consent. MRI including (native) T1 mapping sequences was used to evaluate 145 consecutive patients examined in clinical routine for suspicion of cardiomyopathy. Patients with overt signs of heart failure, i.e. LGE pattern (observed 10–15 min after 0.2 mmol/kg of gadoterate meglumine: Dotarem, Guerbet), irregular heartbeat or myocardial wall, and cavum thickening, were excluded. The remaining 57 patients were divided into two groups with either normal or abnormal functional heart magnetic resonance (MR) parameters. Normal heart function was defined as three of four MR parameters (i.e. left ventricle (LV) end-diastolic volume, LV end-systolic volume, stroke volume, and ejection fraction) being within the normal MR parameter ranges and the fourth still within the border line of normality as defined by Kawel-Boehm et al [25].
Cardiac magnetic resonance imaging
All MR scans were performed on a 1.5-T whole-body scanner (Aera, Siemens Medical Solutions). Functional heart MR parameters were acquired by performing cine imaging steady-state free precession images with echo time (TE) 1.1 ms, repetition time (TR) 42.1 ms, flip angle (FA) 56°, reconstructed voxel size 1.82 × 1.82 × 8 mm, field of view (FOV) 349 × 349, matrix 192 × 192, pixel bandwidth 930 Hz, phase resolution sampling 70%, phase FOV 100%, and GeneRalized Autocalibrating Partial Parallel Acquisition (GRAPPA) acceleration factor 2.
Modified Look-Locker inversion recovery (MOLLI) was implemented in a single breath hold at late diastole, using vendor-provided motion correction for T1 mapping based on image registration with synthetic image estimation [26]. The 5(3)3 MOLLI protocol acquired 5 images after the first inversion pulse, followed by a pause of 3 heartbeats prior to the acquisition of the next 3 images after the second inversion pulse. The protocol’s initial inversion time (TI) was 100 ms, TE 1.12 ms, TR 280.56 ms, and FA 35°. Reconstructed voxel size was 1.41 × 1.41 × 8 mm, FOV 306 × 360, matrix 218 × 256, phase resolution sampling 66%, phase FOV 85%, and GRAPPA acceleration factor 2.
Image analysis
T1 maps were generated by custom-written software (developed in MATLAB version 7.14, The MathWorks) at three short-axis locations (apical, mid-ventricular, and basal) using pixel-wise fitting of a three-parameter model [20]:
to acquire T1 as:
where SI and TI are signal intensity and inversion time, respectively, while A and B are constant values. Two cardiac radiologists (with 5 and 7 years of experience, respectively) and two non-cardiac experts (a radiology technician with 15 years of experience and a non-cardiac radiologist with less than 1 year of experience in cardiac imaging) manually drew LV endocardial and epicardial contours once on the T1 map while carefully avoiding LV blood pool and epicardial fat (Fig. 1). Segmental T1 analysis was conducted on all pixels (without applying endocardial/epicardial inset correction) according to the American Heart Association (AHA) 16-segment model [19] on global myocardium by averaging the 16 segments, different slice locations, and different coronary artery territories [27].
The volumetric cardiac MR parameters were evaluated by a cardiac imaging post-processing radiology technician using QMASS software (Medis Medical Imaging Systems) and checked by a cardiac radiologist (Table 1).
Statistical analysis
Statistical normality testing of data distribution was assessed using the Shapiro-Wilk test using custom-written software (developed in MATLAB version 7.14) [28]. Cardiac MR parameter of a dichotomous variable was compared using the chi-square test and continuous variables were compared using independent t test or the Mann-Whitney U test as appropriate. On normal and abnormal heart function patient groups, each segment T1 quantification was reported both using mean ± standard deviation (SD) and median ± median absolute deviation (MAD) [29, 30] regardless of segment’s statistical normality status. On segments having normally distributed and non-normally distributed pixel-wise T1, comparison between mean and median T1 quantification was assessed by the Mann-Whitney U test. The agreements between mean and median segmental T1 quantification were assessed using the Bland-Altman plot with a limit of agreement (LoA) set to be 1.96 × SD of the difference.
A coefficient of variance (CoV) of the T1 relaxation time was calculated as the SD of the difference divided by the mean and expressed in percentage. Comparison of T1 values between two patient groups on different LV regions was conducted using the independent sample t test for data evaluated by the mean and the Mann-Whitney U test for data evaluated by the median. Multiple comparisons across myocardial regions were done by the Kruskal-Wallis test with the Dunn-Bonferroni post hoc test adjustment. Statistical analyses were performed using IBM SPSS statistics software version 23 (IBM Corporation) with p < 0.05 considered as statistically significant.
Results
Patient classification
According to the criteria of Kawel-Boehm et al [20], 26 of 57 patients were classified in normal heart function group and the remaining 31 patients were classified in abnormal heart function group with similar general characteristics, such as age, heart rate, body surface area, and body mass index (p > 0.05). Their characteristics are listed in Table 1 (and differentiated by gender, in Supplementary Table S1).
Statistical normality of native T1 data distribution
The assessment of AHA 16 segments of LV myocardium from 26 normal patients and 31 abnormal heart function patients resulted in a total of 416 and 496 segments, respectively. With four observers assessing these segments, we obtained 1664 and 1984 segments, respectively.
In all segments of normal patients, statistical normality testing of pixel-wise native T1 per segment showed that 964 of 1664 segments (58%) were statistically non-normally distributed, whereas in all segments of abnormal patients, this statistical distribution was found in 1140 of 1984 segments (57%). In segments having statistically normally distributed pixel-wise T1 (subject for mean quantification), segmental T1 quantification by either mean or median showed no significant difference of T1 value in normal heart function group (p = 0.532) and in abnormal heart function group (p = 0.628). This indicates that in statistically normally distributed data, median quantification is equivalent to the use of the mean. For segments with non-normally distributed pixel-wise T1 (subject for median quantification), a significant difference was found between the two T1 quantifications in both normal (p < 0.001) and abnormal heart (p = 0.003) function groups. This finding indicates that mean quantification cannot be used for statistical non-normally distributed data.
The Bland-Altman plot confirms these claims in normal heart function patients by showing smaller differences of pixel-wise T1 assessed by mean and median quantification for segments having statistically normally distributed pixel-wise T1 (mean difference of 0.95 ms, CoV of 0.85%, and LoA of 15.96 ms) (Fig. 2a) compared with segments with non-normally distributed T1 (mean difference of 9.67 ms, CoV of 1.84%, and LoA of 34.72 ms) (Fig. 2b). Likewise, in abnormal heart function patients (Fig. 2c), pixel-wise T1 had similar smaller Bland-Altman mean difference of 1.04 ms, CoV of 0.78%, and LoA of 14.83 ms in statistically normally distributed data as opposed to higher Bland-Altman of (mean differences of 7.11 ms, CoV of 1.74%, and LoA of 33.39 ms) in non-normally distributed data (Fig. 2d).
Bland-Altman plot assessment of pixel-wise native T1 agreement per segment quantified by means and medians. a Quantification, in normal heart function patients, for segments having statistical normally distributed and statistical non-normally distributed pixel-wise T1 (b). c Quantification, in abnormal heart function patients, for segments having statistical normally distributed and statistical non-normally distributed pixel-wise T1 (d)
Regional T1 analysis and heart function abnormality
In a regional myocardial analysis (Table 2), improvement of interobserver reproducibility of segmental T1 values was found for most regional myocardium areas in normal and abnormal heart function patients when using median compared with the mean for its pixel-wise quantification. This was indicated by CoV reductions, whereas results were similar for observers with different cardiac imaging expertise background (Supplementary Table S2).
Regional T1 analysis of four observers on different LV myocardial regions by using mean and median T1 quantification is presented in Tables 3 and 4, respectively. For each table, the statistical normality testing of its data distribution per LV myocardial region is presented by Supplementary Table S3 for native T1 quantified by its mean and by Supplementary Table S4 for native T1 quantified by its median. Tables S3 and S4 show that most of the T1 data from different myocardial regions are statistically non-normally distributed reflecting inadequate use of mean quantification in Table 3 to differentiate two different patient groups. As a result, the differentiation of T1 values between normal and abnormal heart function groups is undetected in ten of 16 AHA segments of Table 3 (p = 0.059–0.879). When comparing the two patient groups using median quantification (Table 4), significant increase of T1 values is identified in abnormal heart function patients compared with normal heart function in all myocardial regions (p < 0.001–0.024) with exception in the mid-ventricular anteroseptal (p = 0.110) and basal anterior segments (p = 0.080). Heart function differentiation between the two patient groups is thus concluded to be significant in 14 myocardial segments (p < 0.001–0.040) by median quantification compared with only six (p < 0.001–0.042) when using the mean.
Using median quantification, regional LV T1 value in normal heart function patient group was found to be significantly different in the three short-axis slices and in the three coronary artery territories attributed to the 16 AHA segments (Fig. 3a) (p < 0.001–p = 0.023). However, in abnormal heart function patient group (Fig. 3b), T1 value between apical vs. mid-ventricular short-axis slices and between left anterior descending (LAD) and left circumflex artery (LCx) coronary artery territories were not significantly different (p > 0.999 and p = 0.053, respectively).
Discussion
This study shows that median value quantification can be used for segmental native T1 assessment regardless of the distribution of pixel values and therefore can replace mean value quantification where statistical data distribution is normal. Median quantification also showed robustness regardless of the observer’s background by improving interobserver reproducibility of segmental native T1. The superiority of median T1 pixel value quantification compared with mean quantification is confirmed by better differentiation observed between patients with normal and abnormal heart function, especially in the septal regions that are least sensitive to susceptibility artefacts [31]. Therefore, median quantification would be a solution to reduce the influence of any unwanted outlier pixel-wise T1 values. Another study has already promoted MAD of fitting residuals to avoid outliers in T1 fitting process yielding a robust measurement of native T1 [32].
In providing early indication of cardiomyopathy disease in patients with normal cardiac MR functional parameters, native T1 showed no value according to several studies [11, 13, 15]. Our own results obtained with statistical parametric testing and (suboptimal) mean quantification also failed to differentiate between normal and abnormal heart function patients in LV segmental native T1 evaluation. In this study, however, significant increases of T1 values in abnormal heart function patients were found when using median T1 quantification with non-parametric testing instead. Our results also suggest that parametric testing must be performed in native T1 quantification to make sure of statistical normality of the pixel-wise native T1 distribution prior to using means. Alternatively, one can simply use non-parametric testing and medians (as in this study) for the investigation of patient heart condition.
Novel findings in this study of native T1 in normal heart function patients quantified by the medians in different myocardial coronary perfusion territories (i.e. LAD, right coronary artery, LCx, apical, mid-ventricular, and basal), different short-axis slices, and different AHA segments elaborate on those in smaller studies of healthy subjects [16, 19]. The observed variation of T1 value in the LV of normal heart function patients can provide regional baseline T1 values for early detection of diffuse fibrosis and infarct identification.
Suggested elsewhere [33,34,35,36], heart wall T1 elevation is related to coronary microvascular dysfunction (CMD). Camici et al [36] explained that morphological changes of CMD in the absence of myocardial diseases are characterised by microvascular remodelling, endothelial dysfunction, and smooth muscle dysfunction. In patients developing hypertrophic cardiomyopathy, remodelling of intramural coronary arterioles will result in medial and intimal wall thickening [36]. This study reported the elevation of native T1 values in different LV regions of abnormal functional heart patients. Moreover, the variation of native T1 value observed in normal patients between LAD and LCx coronary artery territories was absent in abnormal function heart patients, an observation that might indicate early progression of CMD. But to validate this relationship, more invasive and non-invasive clinical assessment is needed and therefore recommended for further study.
Limitations of this study are that it is retrospective and that patient separation into those having a normal heart function and those without a normal heart function was based on the cardiac MR functional parameters defined by thresholds of just one reported study [25], being, however, very similar to those reported elsewhere [5, 8,9,10,11,12, 14, 15, 37,38,39]. The advice of some [17, 19, 40, 41], to correct native T1 for blood pool, heart rate, age, and gender were not followed through in this study due to the low correlation of T1 with any of these factors. Furthermore, the changes in native T1 values after correction were small and population-dependent (results not shown). Moreover, previous studies reported conflicting findings with regard to these factors’ influence on native T1 value [10, 11, 18, 39, 42]. The T1 maps generated by custom-written software yielded slightly lower values with reduced deviations for all AHA segments compared with the values produced by the Siemens Solution T1 maps (Supplementary Table S5). Investigations into T1 value differences amongst different mapping procedures and into alternative calculation algorithms to improve T1 fitting accuracy [e.g. 43] were not conducted, considered beyond the scope of this study.
Some studies reported the association between diabetes mellitus and the progression of CMD [36, 44, 45]. Another limitation of this study is that diabetes mellitus status of the patients was not recorded.
In conclusion, T1 assessment by observations of medians showed higher interobserver reproducibility compared with mean T1, regardless of statistical normality of data. Increased robustness of myocardial native T1 assessed by pixel-wise medians thus facilitates the early detection of heart function impairment and of differences between LV segments and between the different coronary artery territories.
Abbreviations
- AHA:
-
American Heart Association
- CMD:
-
Coronary microvascular dysfunction
- CoV:
-
Coefficient of variance
- FA:
-
Flip angle
- FOV:
-
Field of view
- GRAPPA:
-
Generalized autocalibrating partial parallel acquisition
- LAD:
-
Left anterior descending
- LCx:
-
Left circumflex artery
- LGE:
-
Late gadolinium enhancement
- LoA:
-
Limit of agreement
- LV:
-
Left ventricle
- MAD:
-
Median absolute deviation
- MOLLI:
-
Modified Look-Locker inversion recovery
- MR:
-
Magnetic resonance
- MRI:
-
Magnetic resonance imaging
- SD:
-
Standard deviation
- TE:
-
Echo time
- TR:
-
Repetition time
References
Puntmann VO, Peker E, Chandrashekhar Y, Nagel E (2016) T1 mapping in characterizing myocardial disease. Circ Res 119:277–299. https://doi.org/10.1161/CIRCRESAHA.116.307974
Raman FS, Kawel-Boehm N, Gai N et al (2013) Modified look-locker inversion recovery T1 mapping indices: assessment of accuracy and reproducibility between magnetic resonance scanners. J Cardiovasc Magn Reson 15:64. https://doi.org/10.1186/1532-429X-15-64
Roujol S, Weingärtner S, Foppa M et al (2014) Accuracy, precision, and reproducibility of four T1 mapping sequences: a head-to-head comparison of MOLLI, ShMOLLI, SASHA, and SAPPHIRE. Radiology 272:683–689. https://doi.org/10.1148/radiol.14140296
Weingärtner S, Meßner NM, Budjan J et al (2016) Myocardial T1-mapping at 3T using saturation-recovery: reference values, precision and comparison with MOLLI. J Cardiovasc Magn Reson 18:84. https://doi.org/10.1186/s12968-016-0302-x
Teixeira T, Hafyane T, Stikov N, Akdeniz C, Greiser A, Friedrich MG (2016) Comparison of different cardiovascular magnetic resonance sequences for native myocardial T1 mapping at 3T. J Cardiovasc Magn Reson 18:65. https://doi.org/10.1186/s12968-016-0286-6
Kellman P, Hansen MS (2014) T1-mapping in the heart: accuracy and precision. J Cardiovasc Magn Reson 16:2. https://doi.org/10.1186/1532-429X-16-2
Olivieri LJ, Kellman P, McCarter RJ, Cross RR, Hansen MS, Spurney CF (2016) Native T1 values identify myocardial changes and stratify disease severity in patients with Duchenne muscular dystrophy. J Cardiovasc Magn Reson 18:72. https://doi.org/10.1186/s12968-016-0292-8
Rogers T, Dabir D, Mahmoud I et al (2013) Standardization of T1 measurements with MOLLI in differentiation between health and disease - the ConSept study. J Cardiovasc Magn Reson 15:78. https://doi.org/10.1186/1532-429X-15-78
Mordi I, Carrick D, Bezerra H, Tzemos N (2016) T1 and T2 mapping for early diagnosis of dilated non-ischaemic cardiomyopathy in middle-aged patients and differentiation from normal physiological adaptation. Eur Heart J Cardiovasc Imaging 17:797–803. https://doi.org/10.1093/ehjci/jev216
Soslow JH, Damon SM, Crum K et al (2016) Increased myocardial native T1 and extracellular volume in patients with Duchenne muscular dystrophy. J Cardiovasc Magn Reson 18:5. https://doi.org/10.1186/s12968-016-0224-7
Dabir D, Child N, Kalra A et al (2014) Reference values for healthy human myocardium using a T1 mapping methodology: results from the international T1 multicenter cardiovascular magnetic resonance study. J Cardiovasc Magn Reson 16:69. https://doi.org/10.1186/s12968-014-0069-x
Pica S, Sado DM, Maestrini V et al (2014) Reproducibility of native myocardial T1 mapping in the assessment of Fabry disease and its role in early detection of cardiac involvement by cardiovascular magnetic resonance. J Cardiovasc Magn Reson 16:99. https://doi.org/10.1186/s12968-014-0099-4
Goebel J, Seifert I, Nensa F et al (2016) Can native T1 mapping differentiate between healthy and diffuse diseased myocardium in clinical routine cardiac MR imaging? PLoS One 11:e0155591. https://doi.org/10.1371/journal.pone.0155591
Yu L, Sun J, Sun J et al (2018) Early detection of myocardial involvement by T 1 mapping of cardiac MRI in idiopathic inflammatory myopathy. J Magn Reson Imaging 48:415–422. https://doi.org/10.1002/jmri.25945
Treibel TA, Zemrak F, Sado DM et al (2015) Extracellular volume quantification in isolated hypertension - changes at the detectable limits? J Cardiovasc Magn Reson 17:74. https://doi.org/10.1186/s12968-015-0176-3
Piechnik SK, Ferreira VM, Dall’Armellina E et al (2010) Shortened modified Look-Locker inversion recovery (ShMOLLI) for clinical myocardial T1-mapping at 1.5 and 3 T within a 9 heartbeat breathhold. J Cardiovasc Magn Reson 12:69. https://doi.org/10.1186/1532-429X-12-69
Messroghli DR, Plein S, Higgins DM et al (2006) Human myocardium: single-breath-hold MR T1 mapping with high spatial resolution--reproducibility study. Radiology 238:1004–1012. https://doi.org/10.1148/radiol.2382041903
Chin CW, Semple S, Malley T et al (2014) Optimization and comparison of myocardial T1 techniques at 3T in patients with aortic stenosis. Eur Heart J Cardiovasc Imaging 15:556–565. https://doi.org/10.1093/ehjci/jet245
Reiter U, Reiter G, Dorr K, Greiser A, Maderthaner R, Fuchsjäger M (2014) Normal diastolic and systolic myocardial T1 values at 1.5-T MR imaging: correlations and blood normalization. Radiology 271:365–372. https://doi.org/10.1148/radiol.13131225
Cameron D, Vassiliou VS, Higgins DM, Gatehouse PD (2018) Towards accurate and precise T 1 and extracellular volume mapping in the myocardium: a guide to current pitfalls and their solutions. MAGMA 31:143–163. https://doi.org/10.1007/s10334-017-0631-2
Saiviroonporn P, Viprakasit V, Boonyasirinant T, Khuhapinant A, Wood JC, Krittayaphong R (2011) Comparison of the region-based and pixel-wise methods for cardiac T2* analysis in 50 transfusion-dependent Thai thalassemia patients. J Comput Assist Tomogr 35:375–381. https://doi.org/10.1097/RCT.0b013e31820eaaf2
Ferguson MR, Otto RK, Bender MA, Kolokythas O, Friedman SD (2013) Liver and heart MR relaxometry in iron loading: reproducibility of three methods. J Magn Reson Imaging 38:987–990. https://doi.org/10.1002/jmri.23937
Positano V, Meloni A, Santarelli MF et al (2015) Fast generation of T2* maps in the entire range of clinical interest: application to thalassemia major patients. Comput Biol Med 56:200–210. https://doi.org/10.1016/j.compbiomed.2014.10.020
Roller FC, Kriechbaum S, Breithecker A et al (2019) Correlation of native T1 mapping with right ventricular function and pulmonary haemodynamics in patients with chronic thromboembolic pulmonary hypertension before and after balloon pulmonary angioplasty. Eur Radiol 29:1565–1573
Kawel-Boehm N, Maceira A, Valsangiacomo-Buechel ER et al (2015) Normal values for cardiovascular magnetic resonance in adults and children. J Cardiovasc Magn Reson 17:29. https://doi.org/10.1186/s12968-015-0111-7
Xue H, Shah S, Greiser A et al (2012) Motion correction for myocardial T1 mapping using image registration with synthetic image estimation. Magn Reson Med 67:1644–1655. https://doi.org/10.1002/mrm.23153
Rinta-Kiikka I, Tuohinen S, Ryymin P et al (2014) Correlation of electrocardiogram and regional cardiac magnetic resonance imaging findings in ST-elevation myocardial infarction: a literature review. Ann Noninvasive Electrocardiol 19:509–523. https://doi.org/10.1111/anec.12210
BenSaida A (2014) Shapiro-Wilk and Shapiro-Francia normality tests. In: MATLAB Cent. File Exch. Available via https://nl.mathworks.com/matlabcentral/fileexchange/13964-shapiro-wilk-and-shapiro-francia-normality-tests. Accessed 24 Mar 2017
Leys C, Ley C, Klein O, Bernarda P, Licata L (2013) Detecting outliers: do not use standard deviation around the mean, use absolute deviation around the median. J Exp Soc Psychol 49:764–766. https://doi.org/10.1016/j.jesp.2013.03.013
Rousseeuw PJ, Croux C (1993) Alternatives to the median absolute deviation. J Am Stat Assoc 88:1273–1283. https://doi.org/10.1080/01621459.1993.10476408
Messroghli DR, Moon JC, Ferreira VM et al (2017) Clinical recommendations for cardiovascular magnetic resonance mapping of T1, T2, T2* and extracellular volume: a consensus statement by the Society for Cardiovascular Magnetic Resonance (SCMR) endorsed by the European Association for Cardiovascular Imaging (EACVI). J Cardiovasc Magn Reson 19:75. https://doi.org/10.1186/s12968-017-0389-8
Kellman P, Arai AE, Xue H (2013) T1 and extracellular volume mapping in the heart: estimation of error maps and the influence of noise on precision. J Cardiovasc Magn Reson 15:56. https://doi.org/10.1186/1532-429X-15-56
Liu A, Wijesurendra RS, Francis JM et al (2016) Adenosine stress and rest T1 mapping can differentiate between ischemic, infarcted, remote, and normal myocardium without the need for gadolinium contrast agents. JACC Cardiovasc Imaging 9:27–36. https://doi.org/10.1016/J.JCMG.2015.08.018
Arnold JR, Karamitsos TD, Bhamra-Ariza P et al (2012) Myocardial oxygenation in coronary artery disease: insights from blood oxygen level–dependent magnetic resonance imaging at 3 tesla. J Am Coll Cardiol 59:1954–1964. https://doi.org/10.1016/J.JACC.2012.01.055
Ferreira VM (2018) T1 mapping of the remote myocardium. J Am Coll Cardiol 71:779–781. https://doi.org/10.1016/j.jacc.2017.12.021
Camici PG, d’Amati G, Rimoldi O (2015) Coronary microvascular dysfunction: mechanisms and functional assessment. Nat Rev Cardiol 12:48–62. https://doi.org/10.1038/nrcardio.2014.160
Fonarow GC, Hsu JJ (2016) Left ventricular ejection fraction: What Is “Normal”? JACC Heart Fail 4:511–513. https://doi.org/10.1016/J.JCHF.2016.03.021
Petersen SE, Aung N, Sanghvi MM et al (2017) Reference ranges for cardiac structure and function using cardiovascular magnetic resonance (CMR) in Caucasians from the UK Biobank population cohort. J Cardiovasc Magn Reson 19:18. https://doi.org/10.1186/s12968-017-0327-9
Liu CY, Liu YC, Wu C et al (2013) Evaluation of age-related interstitial myocardial fibrosis with cardiac magnetic resonance contrast-enhanced T1 mapping: MESA (multi-ethnic study of atherosclerosis). J Am Coll Cardiol 62:1280–1287. https://doi.org/10.1016/j.jacc.2013.05.078
Piechnik SK, Ferreira VM, Lewandowski AJ et al (2013) Normal variation of magnetic resonance T1 relaxation times in the human population at 1.5 T using ShMOLLI. J Cardiovasc Magn Reson 15:13. https://doi.org/10.1186/1532-429X-15-13
Nickander J, Lundin M, Abdula G et al (2017) Blood correction reduces variability and gender differences in native myocardial T1 values at 1.5 T cardiovascular magnetic resonance – a derivation/validation approach. J Cardiovasc Magn Reson 19:41. https://doi.org/10.1186/s12968-017-0353-7
Rauhalammi SMO, Mangion K, Barrientos PH et al (2016) Native myocardial longitudinal ( T1 ) relaxation time: regional, age, and sex associations in the healthy adult heart. J Magn Reson Imaging 44:541–548. https://doi.org/10.1002/jmri.25217
Shao J, Liu D, Sung K, Nguyen KL, Hu P (2017) Accuracy, precision, and reproducibility of myocardial T1 mapping: a comparison of four T1 estimation algorithms for modified look-locker inversion recovery (MOLLI). Magn Reson Med 78:1746–1756. https://doi.org/10.1002/mrm.26565
Crea F, Camici PG, Bairey Merz CN (2014) Coronary microvascular dysfunction: an update. Eur Heart J 35:1101–1111. https://doi.org/10.1093/eurheartj/eht513
Dal Lin C, Tona F, Osto E (2015) Coronary microvascular function and beyond: the crosstalk between hormones, cytokines, and neurotransmitters. Int J Endocrinol 2015:1–17. https://doi.org/10.1155/2015/312848
Acknowledgements
Pandji Triadyaksa receives scholarship from Directorate General of Higher Education of Republic Indonesia (617/E4.4/K/2011).
Funding
The authors state that this work has not received any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is P.E. Sijens, Ph.D.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Study subjects or cohorts overlap
Some study subjects or cohorts have been previously reported in Electronic Presentation Online System (EPOS) European Congress Radiology 2017 Poster No. B-1197.
Methodology
• retrospective
• diagnostic or prognostic study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 56.9 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Triadyaksa, P., Kuijpers, D., Akinci D’Antonoli, T. et al. Early detection of heart function abnormality by native T1: a comparison of two T1 quantification methods. Eur Radiol 30, 652–662 (2020). https://doi.org/10.1007/s00330-019-06364-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-019-06364-9