Abstract
Key message
Extensive leaf transcriptome profiling and differential gene expression analysis of field grown and elicited shoot cultures of L. speciosa suggest that differential synthesis of CRA is mediated primarily by CYP and TS genes, showing functional diversity.
Abstract
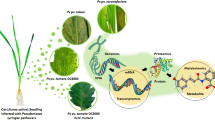
Lagerstroemia speciosa L. is a tree species with medicinal and horticultural attributes. The pentacyclic triterpene, Corosolic acid (CRA) obtained from this species is widely used for the management of diabetes mellitus in traditional medicine. The high mercantile value of the compound and limited availability of innate resources entail exploration of alternative sources for CRA production. Metabolic pathway engineering for enhanced bioproduction of plant secondary metabolites is an attractive proposition for which, candidate genes in the pathway need to be identified and characterized. Therefore, in the present investigation, we focused on the identification of cytochrome P450 (CYP450) and oxidosqualene cyclases (OSC) genes and their differential expression during biosynthesis of CRA. The pattern of differential expression of these genes in the shoot cultures of L. speciosa, elicited with different epigenetic modifiers (azacytidine (AzaC), sodium butyrate (NaBu) and anacardic acid (AA)), was studied in comparison with field grown plant. Further, in vitro cultures with varying (low to high) concentrations of CRA were systematically assessed for the expression of CYP-TS and associated genes involved in CRA biosynthesis by transcriptome sequencing. The sequenced samples were de novo assembled into 180,290 transcripts of which, 92,983 transcripts were further annotated by UniProt. The results are collectively given in co-occurrence heat maps to identify the differentially expressed genes. The combined transcript and metabolite profiles along with RT-qPCR analysis resulted in the identification of CYP-TS genes with high sequence variation. Further, instances of concordant/discordant relation between CRA biosynthesis and CYP-TS gene expression were observed, indicating functional diversity in genes.










Similar content being viewed by others
Data availability
The data underlying this article are provided as supplemental figures and tables in the paper and are also available on request.
Code availability
Not applicable.
Abbreviations
- AA:
-
Anacardic acid
- AzaC:
-
Azacytidine
- BAP:
-
6-Benzylaminopurine
- CYP450:
-
Cytochrome P450
- GO:
-
Gene ontology
- HPLC:
-
High-performance liquid chromatography
- NaBu:
-
Sodium butyrate
- OSC:
-
Oxidosqualene cyclase
- TPS:
-
Terpene synthase
- TS:
-
Terpenoid synthase
References
Abe I, Rohmer M, Prestwich GD (1993) Enzymatic cyclization of squalene and oxidosqualene to sterols and triterpenes. Chem Rev 93:2189–2206. https://doi.org/10.1021/cr00022a009
Ahmad N, Li T, Liu Y, Hoang NQV, Ma X, Zhang X, Liu J, Yao N, Liu X, Li H (2020) Molecular and biochemical rhythms in dihydroflavonol 4-reductase- mediated regulation of leucoanthocyanidin biosynthesis in Carthamus tinctorius L. Ind Crops Prod 156:112838. https://doi.org/10.1016/j.indcrop.2020.112838
Ahn M-A, Lee J, Hyun TK (2023) Histone deacetylase inhibitor, sodium butyrate-induced metabolic modulation in Platycodon grandiflorus roots enhances anti-melanogenic properties. Int J Mol Sci 24:11804. https://doi.org/10.3390/ijms241411804
Alami MM, Ouyang Z, Zhang Y, Shu S, Yang G, Mei Z, Wang X (2022) The current developments in medicinal plant genomics enabled the diversification of secondary metabolites’ biosynthesis. Int J Mol Sci 23:15932. https://doi.org/10.3390/ijms232415932
Alsoufi ASM, Staśkiewicz K, Markowski M (2021) Alterations in oleanolic acid and sterol content in marigold (Calendula officinalis) hairy root cultures in response to stimulation by selected phytohormones. Acta Physiol Plant 43:44. https://doi.org/10.1007/s11738-021-03212-6
Bach TJ (1995) Some new aspects of isoprenoid biosynthesis in plants–a review. Lipids 30:191–202. https://doi.org/10.1007/BF02537822
Banerjee A, Hamberger B (2018) P450s controlling metabolic bifurcations in plant terpene specialized metabolism. Phytochem Rev 17:81–111. https://doi.org/10.1007/s11101-017-9530-4
Bharadwaj R, Kumar SR, Sharma A, Sathishkumar R (2021) Plant metabolic gene clusters: evolution, organization, and their applications in synthetic biology. Front Plant Sci 12. https://doi.org/10.3389/fpls.2021.697318
Bharatiya P, Rathod P, Hiray A, Kate AS (2021) Multifarious elicitors: invoking biosynthesis of various bioactive secondary metabolite in fungi. Appl Biochem Biotechnol 193:668–686. https://doi.org/10.1007/s12010-020-03423-6
Boutanaev AM, Moses T, Zi J, Nelson DR, Mugford ST, Peters RJ, Osbourn A (2015) Investigation of terpene diversification across multiple sequenced plant genomes. Proc Natl Acad Sci 112:E81–E88. https://doi.org/10.1073/pnas.1419547112
Busta L, Serra O, Kim OT, Molinas M, Peré-Fossoul I, Figueras M, Jetter R (2020) Oxidosqualene cyclases involved in the biosynthesis of triterpenoids in Quercus suber cork. Sci Rep 10. https://doi.org/10.1038/s41598-020-64913-5
Chakraborty P (2018) Herbal genomics as tools for dissecting new metabolic pathways of unexplored medicinal plants and drug discovery. Biochimie Open 6:9–16. https://doi.org/10.1016/j.biopen.2017.12.003
Chan EWC, Tan LN, Wong SK (2014) Phytochemistry and pharmacology of Lagerstroemia speciosa: a natural remedy for diabetes. Int J Herb Med 6(2):81–87
Chen X, Luck K, Rabe P, Dinh CQ, Shaulsky G, Nelson DR, Gershenzon J, Dickschat JS, Köllner TG, Chen F (2019) A terpene synthase-cytochrome P450 cluster in Dictyostelium discoideum produces a novel trisnorsesquiterpene. eLife 8: e44352. https://doi.org/10.7554/eLife.44352
Cherukupalli N, Divate M, Mittapelli SR, Khareedu VR, Vudem DR (2016) De Novo assembly of leaf transcriptome in the medicinal plant Andrographis Paniculata. Front Plant Sci 7. https://doi.org/10.3389/fpls.2016.01203.
Cho HJ, Kim SY, Kim KH, Kang WK, Kim JI, Oh ST, Kim JS, An CH (2009) The combination effect of sodium butyrate and 5-Aza-2’-deoxycytidine on radiosensitivity in RKO colorectal cancer and MCF-7 breast cancer cell lines. World J Surg Oncol 7:49. https://doi.org/10.1186/1477-7819-7-49
Christianson DW (2017) Structural and chemical biology of Terpenoid Cyclases. Chem Rev 117:11570–11648. https://doi.org/10.1021/acs.chemrev.7b00287
Christman JK (2002) 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 21:5483–5495. https://doi.org/10.1038/sj.onc.1205699
Dixon RA (1999) Plant natural products: the molecular genetic basis of biosynthetic diversity. Curr Opin Biotechnol 10:192–197. https://doi.org/10.1016/S0958-1669(99)80034-2
Dong L, Pollier J, Bassard J-E, Ntallas G, Almeida A, Lazaridi E, Khakimov B, Arendt P, de Oliveira LS, Lota F, Goossens A, Michoux F, Bak S (2018) Co-expression of squalene epoxidases with triterpene cyclases boosts production of triterpenoids in plants and yeast. Metab Eng 49:1–12. https://doi.org/10.1016/j.ymben.2018.07.002
Eisenreich W, Schwarz M, Cartayrade A, Arigoni D, Zenk MH, Bacher A (1998) The deoxyxylulose phosphate pathway of terpenoid biosynthesis in plants and microorganisms. Chem Biol 5:R221-233. https://doi.org/10.1016/s1074-5521(98)90002-3
Fukushima EO, Seki H, Ohyama K, Ono E, Umemoto N, Mizutani M, Saito K, Muranaka T (2011) CYP716A subfamily members are multifunctional oxidases in triterpenoid biosynthesis. Plant Cell Physiol 52:2050–2061. https://doi.org/10.1093/pcp/pcr146
Ghosh S (2016) Biosynthesis of structurally diverse triterpenes in plants: the role of Oxidosqualene Cyclases. Proc Indian Natl Sci Acad 82:1189–1210. https://doi.org/10.16943/ptinsa/2016/48578
Ghosh S (2017) Triterpene structural diversification by plant cytochrome P450 enzymes. Front Plant Sci 8:1886. https://doi.org/10.3389/fpls.2017.01886
GlucoFit 18% Corosolic Acid [WWW Document] (n.d.) https://www.dclabs.com/glucotrim
Grassi G, Maccaroni P, Meyer R, Kaiser H, D’Ambrosio E, Pascale E, Grassi M, Kuhn A, Di Nardo P, Kandolf R, Küpper J-H (2003) Inhibitors of DNA methylation and histone deacetylation activate cytomegalovirus promoter-controlled reporter gene expression in human glioblastoma cell line U87. Carcinogenesis 24:1625–1635. https://doi.org/10.1093/carcin/bgg118
Hou W, Li Y, Zhang Q, Wei X, Peng A, Chen L, Wei Y (2009) Triterpene acids isolated from Lagerstroemia speciosa leaves as α-glucosidase inhibitors. Phytother Res 23:614–618. https://doi.org/10.1002/ptr.2661
Iturbe-Ormaetxe I, Haralampidis K, Papadopoulou K, Osbourn AE (2003) Molecular cloning and characterization of triterpene synthases from Medicago truncatula and Lotus japonicus. Plant Mol Biol 51:731–743. https://doi.org/10.1023/a:1022519709298
Jayakumar KS, Sajan JS, Aswati Nair R, Padmesh Pillai P, Deepu S, Padmaja R, Agarwal A, Pandurangan AG (2014) Corosolic acid content and SSR markers in Lagerstroemia speciosa (L.) Pers.: a comparative analysis among populations across the Southern Western Ghats of India. Phytochemistry 106:94–103. https://doi.org/10.1016/j.phytochem.2014.07.004
Judy WV, Hari SP, Stogsdill WW, Judy JS, Naguib YMA, Passwater R (2003) Antidiabetic activity of a standardized extract (Glucosol) from Lagerstroemia speciosa leaves in Type II diabetics A Dose-Dependence Study. J Ethnopharmacol 87:115–117. https://doi.org/10.1016/s0378-8741(03)00122-3
Karunanithi PS, Zerbe P (2019) Terpene synthases as metabolic gatekeepers in the evolution of plant terpenoid chemical diversity. Front Plant Sci 10:1166. https://doi.org/10.3389/fpls.2019.01166
Lee MH, Lee J, Choi SH, Jie EY, Jeong JC, Kim CY, Kim SW (2020) The effect of sodium butyrate on adventitious shoot formation varies among the plant species and the explant types. Int J Mol Sci 21:E8451. https://doi.org/10.3390/ijms21228451
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. https://doi.org/10.1186/1471-2105-12-323
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Mafezoli J, Xu Y, Hilário F, Freidhof B, Espinosa-Artiles P, dos Santos LC, de Oliveira MCF, Gunatilaka AAL (2018) Modulation of polyketide biosynthetic pathway of the endophytic fungus, Anteaglonium sp. FL0768, by copper (II) and anacardic acid. Phytochem Lett 28:157–163. https://doi.org/10.1016/j.phytol.2018.10.011
Malhotra K, Franke J (2022) Cytochrome P450 monooxygenase-mediated tailoring of triterpenoids and steroids in plants. Beilstein J Org Chem 18:1289–1310. https://doi.org/10.3762/bjoc.18.135
McGarvey DJ, Crotea R (1995) Terpenoid metabolism. Plant Cell 7:1015–1026. https://doi.org/10.1105/tpc.7.7.1015
Miettinen K, Pollier J, Buyst D, Arendt P, Csuk R, Sommerwerk S, Moses T, Mertens J, Sonawane PD, Pauwels L, Aharoni A, Martins J, Nelson DR, Goossens A (2017) The ancient CYP716 family is a major contributor to the diversification of eudicot triterpenoid biosynthesis. Nat Commun 8. https://doi.org/10.1038/ncomms14153
Nakamura M, Linh TM, Lien LQ, Suzuki H, Mai NC, Giang VH, Tamura K, Thanh NV, Suzuki H, Misaki R, Muranaka T, Ban NK, Fujiyama K, Seki H (2018) Transcriptome sequencing and identification of cytochrome P450 monooxygenases involved in the biosynthesis of maslinic acid and corosolic acid in Avicennia marina. Plant Biotechnol (tokyo) 35:341–348. https://doi.org/10.5511/plantbiotechnology.18.0810a
Nazaruk J, Borzym-Kluczyk M (2015) The role of triterpenes in the management of diabetes mellitus and its complications. Phytochem Rev 14:675–690. https://doi.org/10.1007/s11101-014-9369-x
Nelson DR (2009) The cytochrome P450 homepage. Hum Genomics 4:59. https://doi.org/10.1186/1479-7364-4-1-59
Nelson D, Werck-Reichhart D (2011) A P450-centric view of plant evolution. Plant J 66:194–211. https://doi.org/10.1111/j.1365-313X.2011.04529.x
Nelson AT, Camelio AM, Claussen KR, Cho J, Tremmel L, DiGiovanni J, Siegel D (2015) Synthesis of oxygenated oleanolic and ursolic acid derivatives with anti-inflammatory properties. Bioorg Med Chem Lett 25:4342–4346. https://doi.org/10.1016/j.bmcl.2015.07.029
Nes WR, McKean ML (1977) Biochemistry of steroids and other isoprenoids. University Park Press, Baltimore
Oboh M, Govender L, Siwela M, Mkhwanazi BN (2021) Anti-diabetic potential of plant-based pentacyclic triterpene derivatives: progress made to improve efficacy and bioavailability. Molecules 26:7243. https://doi.org/10.3390/molecules26237243
Ozyigit II, Dogan I, Hocaoglu-Ozyigit A, Yalcin B, Erdogan A, Yalcin IE, Cabi E, Kaya Y (2023) Production of secondary metabolites using tissue culture-based biotechnological applications. Front Plant Sci 14:1132555. https://doi.org/10.3389/fpls.2023.1132555
Pateraki I, Heskes A, Hamberger B (2015) Cytochromes P450 for terpene functionalisation and metabolic engineering. Adv Biochem Eng Biotechnol 148. https://doi.org/10.1007/10_2014_301
Prism 10.0.2 Release Notes [WWW Document] (n.d.) https://www.graphpad.com/updates/prism-1002-release-notes
QIAGEN (n.d.) RNeasy Plant Mini Kit | Plant RNA Extraction | QIAGEN [WWW Document]. https://www.qiagen.com/us/products/discovery-and-translational-research/dna-rna-purification/rna-purification/total-rna/rneasy-plant-mini-kit. Accessed 26 Feb 2024
Rahman M, Amin MN, Rahman M, Sharmin R (2010) In vitro adventitious shoot organogenesis and plantlet regeneration from leaf-derived callus of Lagerstroemia speciosa (L.) Pers. Propag Ornamental Plants 10:149–155
Rohmer M, Knani M, Simonin P, Sutter B, Sahm H (1993) Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem J 295:517–524. https://doi.org/10.1042/bj2950517
Sandeep MRC, Chanotiya CS, Mukhopadhyay P, Ghosh S (2019) Oxidosqualene cyclase and CYP716 enzymes contribute to triterpene structural diversity in the medicinal tree banaba. New Phytol 222:408–424. https://doi.org/10.1111/nph.15606
Schubert M, Lindgreen S, Orlando L (2016) AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res Notes 9:88. https://doi.org/10.1186/s13104-016-1900-2
Seki H, Tamura K, Muranaka T (2015) P450s and UGTs: key players in the structural diversity of Triterpenoid Saponins. Plant Cell Physiol 56:1463–1471. https://doi.org/10.1093/pcp/pcv062
Shi H, Wei SH, Leu Y-W, Rahmatpanah F, Liu JC, Yan PS, Nephew KP, Huang TH-M (2003) Triple analysis of the cancer epigenome: an integrated microarray system for assessing gene expression, DNA methylation, and histone acetylation. Can Res 63:2164–2171
Shim KS, Lee S-U, Ryu SY, Min YK, Kim SH (2009) Corosolic acid stimulates osteoblast differentiation by activating transcription factors and MAP kinases. Phytother Res 23:1754–1758. https://doi.org/10.1002/ptr.284
Sivakumar G, Vail DR, Nair V, Medina-Bolivar F, Lay JO (2009) Plant-based corosolic acid: future anti-diabetic drug? Biotechnol J 4:1704–1711. https://doi.org/10.1002/biot.200900207
Sprenger GA, Schörken U, Wiegert T, Grolle S, de Graaf AA, Taylor SV, Begley TP, Bringer-Meyer S, Sahm H (1997) Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-D-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc Natl Acad Sci USA 94:12857–12862. https://doi.org/10.1073/pnas.94.24.12857
Stohs SJ, Miller H, Kaats GR (2012) A review of the efficacy and safety of banaba (Lagerstroemia speciosa L.) and corosolic acid. Phytother Res 26:317–324. https://doi.org/10.1002/ptr.3664
Su W, Jing Y, Lin S Yue Z, Yang X, Xu J, Wu J, Zhang Z, Xia R, Zhu J, An N, Chen H, Hong Y, Yuan Y, Long T, Zhang L, Jiang Y, Liu Z, Zhang H, Gao Y, Liu Y, Lin H, Wang H, Yant L, Lin S, Liu Z (2021) Polyploidy underlies co-option and diversification of biosynthetic triterpene pathways in the apple tribe. Proc Natl Acad Sci USA 118. https://doi.org/10.1073/pnas.2101767118
Takayama H, Kitajima M, Ishizuka T, Seo S (2006) Process for producing corosolic acid. US7071229B2
ThermoFisher (n.d.) Verso cDNA Synthesis Kit [WWW Document]. https://www.thermofisher.com/order/catalog/product/AB1453A. Accessed 26 Feb 2024
Vijayan A, Padmesh Pillai P, Hemanthakumar AS, Krishnan PN (2015) Improved in vitro propagation, genetic stability and analysis of corosolic acid synthesis in regenerants of Lagerstroemia speciosa (L.) Pers. by HPLC and gene expression profiles. Plant Cell, Tissue Organ Cult 120:1209–1214. https://doi.org/10.1007/s11240-014-0665-3
Wen X, Xia J, Cheng K, Zhang L, Zhang P, Liu J, Zhang L, Ni P, Sun H (2007) Pentacyclic triterpenes. Part 5: synthesis and SAR study of corosolic acid derivatives as inhibitors of glycogen phosphorylases. Bioorg Med Chem Lett 17:5777–5782. https://doi.org/10.1016/j.bmcl.2007.08.057
Wen X, Sun H, Liu J, Cheng K, Zhang P, Zhang L, Hao J, Zhang L, Ni P, Zographos SE, Leonidas DD, Alexacou K-M, Gimisis T, Hayes JM, Oikonomakos NG (2008) Naturally occurring pentacyclic triterpenes as inhibitors of glycogen phosphorylase: synthesis, structure−activity relationships, and X-ray crystallographic studies. J Med Chem 51:3540–3554. https://doi.org/10.1021/jm8000949
Xu Y, Zhao Y, Xu Y, Guan Y, Zhang X, Chen Y, Wu Q, Zhu G, Chen Y, Sun F, Wang J, Yu Y (2017) Blocking inhibition to YAP by ActinomycinD enhances anti-tumor efficacy of Corosolic acid in treating liver cancer. Cell Signal 29:209–217. https://doi.org/10.1016/j.cellsig.2016.11.001
Yamaguchi Y, Yamada K, Yoshikawa N, Nakamura K, Haginaka J, Kunitomo M (2006) Corosolic acid prevents oxidative stress, inflammation and hypertension in SHR/NDmcr-cp rats, a model of metabolic syndrome. Life Sci 79:2474–2479. https://doi.org/10.1016/j.lfs.2006.08.007
Yang B-C, Lee M-S, Lin M-K, Chang W-T (2022) 5-Azacytidine increases tanshinone production in Salvia miltiorrhiza hairy roots through epigenetic modulation. Sci Rep 12:9349. https://doi.org/10.1038/s41598-022-12577-8
Yang C, Halitschke R, O’Connor SE (2023) OXIDOSQUALENE CYCLASE 1 and 2 influence triterpene biosynthesis and defense in Nicotiana attenuata. Plant Physiol kiad643. https://doi.org/10.1093/plphys/kiad643
Zhao L, Chang W, Xiao Y, Liu Y, Liu H (2013) Methylerythritol phosphate pathway of isoprenoid biosynthesis. Annu Rev Biochem 82:497–530. https://doi.org/10.1146/annurev-biochem-052010-100934
Zhao J, Zhou H, An Y, Shen K, Yu L (2021) Biological effects of corosolic acid as an anti-inflammatory, anti-metabolic syndrome and anti-neoplastic natural compound. Oncol Lett 21:84. https://doi.org/10.3892/ol.2020.12345
Zhou H-C, Shamala LF, Yi X-K, Yan Z, Wei S (2020) Analysis of terpene synthase family genes in Camellia sinensis with an emphasis on abiotic stress conditions. Sci Rep 10:933. https://doi.org/10.1038/s41598-020-57805-1
Acknowledgements
We greatly acknowledge the Vice Chancellor of Central University of Kerala, Kasaragod, for all the supports rendered throughout this study. K.S acknowledges with thanks the Department of Science and Technology, Govt. of India for providing the DST-INSPIRE fellowship (DST/INSPIRE Fellowship/2017/IF170450).
Funding
The work was partly supported by funding from Central University of Kerala.
Author information
Authors and Affiliations
Contributions
K.S performed all analyses and experiments concerning tissue culture, CYP450 enzymes and TS. K.S and S.P conducted the validation studies of transcriptome data. P.P conceptualized the research problem and designed the experiments. K.S prepared the draft and P.P refined and revised the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Communicated by Prakash Lakshmanan.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Surendran, K., Pradeep, S. & Pillai, P.P. Comparative transcriptome and metabolite profiling reveal diverse pattern of CYP-TS gene expression during corosolic acid biosynthesis in Lagerstroemia speciosa (L.) Pers. Plant Cell Rep 43, 122 (2024). https://doi.org/10.1007/s00299-024-03203-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00299-024-03203-0