Abstract
Key message
ZmWRKY64 positively regulates Arabidopsis and maize Cd stress through modulating Cd uptake, translocation, and ROS scavenging genes expression.
Abstract
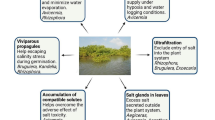
Cadmium (Cd) is a highly toxic heavy metal with severe impacts on crops growth and development. The WRKY transcription factor is a significant regulator influencing plant stress response. Nevertheless, the function of the WRKY protein in maize Cd stress response remains unclear. Here, we identified a maize WRKY gene, ZmWRKY64, the expression of which was enhanced in maize roots and leaves under Cd stress. ZmWRKY64 was localized in the nucleus and displayed transcriptional activity in yeast. Heterologous expression of ZmWRKY64 in Arabidopsis diminished Cd accumulation in plants by negatively regulating the expression of AtIRT1, AtZIP1, AtHMA2, AtNRAMP3, and AtNRAMP4, which are involved in Cd uptake and transport, resulting in Cd stress tolerance. Knockdown of ZmWRKY64 in maize led to excessive Cd accumulation in leaf cells and in the cytosol of the root cells, resulting in a Cd hypersensitive phenotype. Further analysis confirmed that ZmWRKY64 positively regulated ZmABCC4, ZmHMA3, ZmNRAMP5, ZmPIN2, ZmABCG51, ZmABCB13/32, and ZmABCB10, which may influence Cd translocation and auxin transport, thus mitigating Cd toxicity in maize. Moreover, ZmWRKY64 could directly enhance the transcription of ZmSRG7, a reported key gene regulating reactive oxygen species homeostasis under abiotic stress. Our results indicate that ZmWRKY64 is important in maize Cd stress response. This work provides new insights into the WRKY transcription factor regulatory mechanism under a Cd-polluted environment and may lead to the genetic improvement of Cd tolerance in maize.









Similar content being viewed by others
Data availability
The raw sequence data generated by this research have been deposited in the National Center for Biotechnology Information under the accession number PRJNA936919 for RNA sequencing.
References
Aryal B, Huynh J, Schneuwly J, Siffert A, Liu J, Alejandro S, Ludwig-Muller J, Martinoia E, Geisler M (2019) ABCG36/PEN3/PDR8 is an exporter of the auxin precursor, indole-3-butyric acid, and involved in auxin-controlled development. Front Plant Sci 10:899
Brunetti P, Zanella L, De Paolis A, Di Litta D, Cecchetti V, Falasca G, Barbieri M, Altamura MM, Costantino P, Cardarelli M (2015) Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J Exp Bot 66:3815–3829
Cai Z, Xian P, Wang H, Lin R, Lian T, Cheng Y, Ma Q, Nian H (2020) Transcription factor GmWRKY142 confers cadmium resistance by up-regulating the cadmium tolerance 1-like genes. Front Plant Sci 11:724
Cailliatte R, Schikora A, Briat JF, Mari S, Curie C (2010) High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell 22:904–917
Cao Y, Zhao X, Liu Y, Wang Y, Wu W, Jiang Y, Liao C, Xu X, Gao S, Shen Y, Lan H, Zou C, Pan G, Lin H (2019) Genome-wide identification of ZmHMAs and association of natural variation in ZmHMA2 and ZmHMA3 with leaf cadmium accumulation in maize. PeerJ 7:e7877
Chang JD, Huang S, Konishi N, Wang P, Chen J, Huang XY, Ma JF, Zhao FJ (2020) Overexpression of the manganese/cadmium transporter OsNRAMP5 reduces cadmium accumulation in rice grain. J Exp Bot 71:5705–5715
Chao DY, Silva A, Baxter I, Huang YS, Nordborg M, Danku J, Lahner B, Yakubova E, Salt DE (2012) Genome-wide association studies identify heavy metal ATPase3 as the primary determinant of natural variation in leaf cadmium in Arabidopsis thaliana. PLoS Genet 8:e1002923
Chen F, Hu Y, Vannozzi A, Wu KC, Cai HY, Qin Y, Mullis A, Lin ZG, Zhang LS (2017) The WRKY transcription factor family in model plants and crops. Crit Rev Plant Sci 36:311–335
Chen Y, Chao ZF, Jin M, Wang YL, Li Y, Wu JC, Xiao Y, Peng Y, Lv QY, Gui S, Wang X, Han ML, Fernie AR, Chao DY, Yan J (2022) A heavy metal transporter gene ZmHMA3a promises safe agricultural production on cadmium-polluted arable land. J Genet Genom 50:130–134
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Dang F, Lin J, Chen Y, Li GX, Guan D, Zheng SJ, He S (2019) A feedback loop between CaWRKY41 and H2O2 coordinates the response to Ralstonia solanacearum and excess cadmium in pepper. J Exp Bot 70:1581–1595
Fu S, Lu Y, Zhang X, Yang G, Chao D, Wang Z, Shi M, Chen J, Chao DY, Li R, Ma JF, Xia J (2019) The ABC transporter ABCG36 is required for cadmium tolerance in rice. J Exp Bot 70:5909–5918
Gu L, Jiang T, Zhang C, Li X, Wang C, Zhang Y, Li T, Dirk LMA, Downie AB, Zhao T (2019) Maize HSFA2 and HSBP2 antagonistically modulate raffinose biosynthesis and heat tolerance in Arabidopsis. Plant J 100:128–142
Gu L, Chen X, Hou Y, Wang H, Wang H, Zhu B, Du X (2023) ZmWRKY70 activates the expression of hypoxic responsive genes in maize and enhances tolerance to submergence in Arabidopsis. Plant Physiol Biochem 201:107861
Han Y, Fan T, Zhu X, Wu X, Ouyang J, Jiang L, Cao S (2019) WRKY12 represses GSH1 expression to negatively regulate cadmium tolerance in Arabidopsis. Plant Mol Biol 99:149–159
Hu W, Ren Q, Chen Y, Xu G, Qian Y (2021) Genome-wide identification and analysis of WRKY gene family in maize provide insights into regulatory network in response to abiotic stresses. BMC Plant Biol 21:427
Huang L, Liu L, Zhang T, Zhao D, Li H, Sun H, Kinney PL, Pitiranggon M, Chillrud S, Ma LQ, Navas-Acien A, Bi J, Yan B (2019) An interventional study of rice for reducing cadmium exposure in a Chinese industrial town. Environ Int 122:301–309
Hussain D, Haydon MJ, Wang Y, Wong E, Sherson SM, Young J, Camakaris J, Harper JF, Cobbett CS (2004) P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 16:1327–1339
Jenness MK, Tayengwa R, Bate GA, Tapken W, Zhang Y, Pang C, Murphy AS (2022) Loss of multiple ABCB auxin transporters recapitulates the major twisted dwarf 1 phenotypes in Arabidopsis thaliana. Front Plant Sci 13:840260
Jia ZZ, Li MZ, Wang HC, Zhu B, Gu L, Du XY, Ren MJ (2021) TaWRKY70 positively regulates TaCAT5 enhanced Cd tolerance in transgenic Arabidopsis. Environ Exp Bot 190:104591
Jiang Y, Han J, Xue W, Wang J, Wang B, Liu L, Zou J (2021) Overexpression of SmZIP plays important roles in Cd accumulation and translocation, subcellular distribution, and chemical forms in transgenic tobacco under Cd stress. Ecotoxicol Environ Saf 214:112097
Kim DY, Bovet L, Maeshima M, Martinoia E, Lee Y (2007) The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J 50:207–218
Kolahi M, Mohajel Kazemi E, Yazdi M, Goldson-Barnaby A (2020) Oxidative stress induced by cadmium in lettuce (Lactuca sativa Linn.): oxidative stress indicators and prediction of their genes. Plant Physiol Biochem 146:71–89
Lanquar V, Lelièvre F, Barbier-Brygoo H, Thomine S (2004) Regulation and function of AtNRAMP4 metal transporter protein. Soil Sci Plant Nutr 50:1141–1150
Li SS, Wang M, Zhao ZQ, Ma CB, Chen SB (2018) Adsorption and desorption of Cd by soil amendment: mechanisms and environmental implications in field-soil remediation. Sustainability 10:2337
Liu J, Ghelli R, Cardarelli M, Geisler M (2022) Arabidopsis TWISTED DWARF1 regulates stamen elongation by differential activation of ABCB1,19-mediated auxin transport. J Exp Bot 73:4818–4831
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408
Luo JS, Huang J, Zeng DL, Peng JS, Zhang GB, Ma HL, Guan Y, Yi HY, Fu YL, Han B, Lin HX, Qian Q, Gong JM (2018) A defensin-like protein drives cadmium efflux and allocation in rice. Nat Commun 9:645
Lyu M, Liu H, Waititu JK, Sun Y, Wang H, Fu J, Chen Y, Liu J, Ku L, Cheng X (2021) TEAseq-based identification of 35,696 Dissociation insertional mutations facilitates functional genomic studies in maize. J Genet Genom 48:961–971
Ma H, Wei M, Wang Z, Hou S, Li X, Xu H (2020) Bioremediation of cadmium polluted soil using a novel cadmium immobilizing plant growth promotion strain Bacillus sp. TZ5 loaded on biochar. J Hazard Mater 388:122065
Matthes MS, Best NB, Robil JM, Malcomber S, Gallavotti A, McSteen P (2019) Auxin EvoDevo: conservation and diversification of genes regulating auxin biosynthesis, transport, and signaling. Mol Plant 12:298–320
Meng Y, Huang J, Jing H, Wu Q, Shen R, Zhu X (2022) Exogenous abscisic acid alleviates Cd toxicity in Arabidopsis thaliana by inhibiting Cd uptake, translocation and accumulation, and promoting Cd chelation and efflux. Plant Sci 325:111464
Pang K, Li Y, Liu M, Meng Z, Yu Y (2013) Inventory and general analysis of the ATP-binding cassette (ABC) gene superfamily in maize (Zea mays L.). Gene 526:411–428
Park J, Song WY, Ko D, Eom Y, Hansen TH, Schiller M, Lee TG, Martinoia E, Lee Y (2012) The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J 69:278–288
Porebski S, Bailey LG, Baum BR (1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep 15:8–15
Rizwan M, Ali S, Adrees M, Rizvi H, Zia-Ur-Rehman M, Hannan F, Qayyum MF, Hafeez F, Ok YS (2016) Cadmium stress in rice: toxic effects, tolerance mechanisms, and management: a critical review. Environ Sci Pollut Res Int 23:17859–17879
Sasaki A, Yamaji N, Yokosho K, Ma JF (2012) Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 24:2155–2167
Sheng Y, Yan X, Huang Y, Han Y, Zhang C, Ren Y, Fan T, Xiao F, Liu Y, Cao S (2019) The WRKY transcription factor, WRKY13, activates PDR8 expression to positively regulate cadmium tolerance in Arabidopsis. Plant Cell Environ 42:891–903
Sun L, Zhang X, Ouyang W, Yang E, Cao Y, Sun R (2022) Lowered Cd toxicity, uptake and expression of metal transporter genes in maize plant by ACC deaminase-producing bacteria Achromobacter sp. J Hazard Mater 423:127036
Tan L, Qu M, Zhu Y, Peng C, Wang J, Gao D, Chen C (2020) ZINC TRANSPORTER5 and ZINC TRANSPORTER9 function synergistically in zinc/cadmium uptake. Plant Physiol 183:1235–1249
Tang L, Mao B, Li Y, Lv Q, Zhang L, Chen C, He H, Wang W, Zeng X, Shao Y, Pan Y, Hu Y, Peng Y, Fu X, Li H, Xia S, Zhao B (2017) Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci Rep 7:14438
Tang B, Luo M, Zhang Y, Guo H, Li J, Song W, Zhang R, Feng Z, Kong M, Li H, Cao Z, Lu X, Li D, Zhang J, Wang R, Wang Y, Chen Z, Zhao Y, Zhao J (2021) Natural variations in the P-type ATPase heavy metal transporter gene ZmHMA3 control cadmium accumulation in maize grains. J Exp Bot 72:6230–6246
Tang Z, Wang HQ, Chen J, Chang JD, Zhao FJ (2022) Molecular mechanisms underlying the toxicity and detoxification of trace metals and metalloids in plants. J Integr Plant Biol 65:570–593
Tao L, Xiao X, Huang Q, Zhu H, Feng Y, Li Y, Li X, Guo Z, Liu J, Wu F, Pirayesh N, Mahmud S, Shen RF, Shabala S, Baluska F, Shi L, Yu M (2023) Boron supply restores aluminum-blocked auxin transport by the modulation of PIN2 trafficking in the root apical transition zone. Plant J 114:176–192
Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI (2000) Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc Natl Acad Sci U S A 97:4991–4996
Ueno D, Yamaji N, Kono I, Huang CF, Ando T, Yano M, Ma JF (2010) Gene limiting cadmium accumulation in rice. Proc Natl Acad Sci U S A 107:16500–16505
Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14:1223–1233
Wang HQ, Xuan W, Huang XY, Mao C, Zhao FJ (2021) Cadmium inhibits lateral root emergence in rice by disrupting OsPIN-mediated auxin distribution and the protective effect of OsHMA3. Plant Cell Physiol 62:166–177
Wanke D, Kolukisaoglu HU (2010) An update on the ABCC transporter family in plants: many genes, many proteins, but how many functions? Plant Biol (stuttg) 12(Suppl 1):15–25
Wei X, Fan X, Zhang H, Jiao P, Jiang Z, Lu X, Liu S, Guan S, Ma Y (2022) Overexpression of ZmSRG7 improves drought and salt tolerance in maize (Zea mays L.). Int J Mol Sci 23:13349
Wong CKE, Cobbett CS (2009) HMA P-type ATPases are the major mechanism for root-to-shoot Cd translocation in Arabidopsis thaliana. New Phytol 181:71–78
Wu HF, Wang JY, Ou YJ, Li BB, Jiang WS, Liu DH, Zou JH (2017) Characterisation of early responses to cadmium in roots of Salix matsudana Koidz. Toxicol Environ Chem 99:913–925
Yan H, Xu W, Xie J, Gao Y, Wu L, Sun L, Feng L, Chen X, Zhang T, Dai C, Li T, Lin X, Zhang Z, Wang X, Li F, Zhu X, Li J, Li Z, Chen C, Ma M, Zhang H, He Z (2019) Variation of a major facilitator superfamily gene contributes to differential cadmium accumulation between rice subspecies. Nat Commun 10:2562
Yang G, Wang C, Wang Y, Guo Y, Zhao Y, Yang C, Gao C (2016) Overexpression of ThVHAc1 and its potential upstream regulator, ThWRKY7, improved plant tolerance of Cadmium stress. Sci Rep 6:18752
Yang G, Fu S, Huang J, Li L, Long Y, Wei Q, Wang Z, Chen Z, Xia J (2021) The tonoplast-localized transporter OsABCC9 is involved in cadmium tolerance and accumulation in rice. Plant Sci 307:110894
Yang Z, Yang F, Liu JL, Wu HT, Yang H, Shi Y, Liu J, Zhang YF, Luo YR, Chen KM (2022) Heavy metal transporters: functional mechanisms, regulation, and application in phytoremediation. Sci Total Environ 809:151099
Yue R, Tie S, Sun T, Zhang L, Yang Y, Qi J, Yan S, Han X, Wang H, Shen C (2015) Genome-wide identification and expression profiling analysis of ZmPIN, ZmPILS, ZmLAX and ZmABCB auxin transporter gene families in maize (Zea mays L.) under various abiotic stresses. PLoS ONE 10:e0118751
Yue R, Lu C, Qi J, Han X, Yan S, Guo S, Liu L, Fu X, Chen N, Yin H, Chi H, Tie S (2016) Transcriptome analysis of cadmium-treated roots in maize (Zea mays L.). Front Plant Sci 7:1298
Zhang X, Rui H, Zhang F, Hu Z, Xia Y, Shen Z (2018) Overexpression of a functional Vicia sativa PCS1 homolog increases cadmium tolerance and phytochelatins synthesis in arabidopsis. Front Plant Sci 9:107
Zhang L, Wu J, Tang Z, Huang XY, Wang X, Salt DE, Zhao FJ (2019a) Variation in the BrHMA3 coding region controls natural variation in cadmium accumulation in Brassica rapa vegetables. J Exp Bot 70:5865–5878
Zhang W, Song J, Yue S, Duan K, Yang H (2019b) MhMAPK4 from Malus hupehensis Rehd. decreases cell death in tobacco roots by controlling Cd(2+) uptake. Ecotoxicol Environ Saf 168:230–240
Zhang Y, Wang Z, Liu Y, Zhang T, Liu J, You Z, Huang P, Zhang Z, Wang C (2023) Plasma membrane-associated calcium signaling modulates cadmium transport. New Phytol 238:313–331
Zhao X, Luo L, Cao Y, Liu Y, Li Y, Wu W, Lan Y, Jiang Y, Gao S, Zhang Z, Shen Y, Pan G, Lin H (2018) Genome-wide association analysis and QTL mapping reveal the genetic control of cadmium accumulation in maize leaf. BMC Genomics 19:91
Zhao FJ, Tang Z, Song JJ, Huang XY, Wang P (2022) Toxic metals and metalloids: uptake, transport, detoxification, phytoremediation, and crop improvement for safer food. Mol Plant 15:27–44
Acknowledgements
We thank Professor Chen Yan-Hui of the College of Agronomy at Henan Agricultural University for constructing the maize mutant library. Additionally, we extend our gratitude to the Major Platform Center at the Institute of Crop Science, Chinese Academy of Agricultural Sciences, for providing the maize mutants.
Funding
This research was funded by the National Natural Science Foundation of China (grant number 32160434), the Guizhou Provincial Key Technology R&D Program (grant number Qian Kehe Support [2022]key026), the Guizhou Provincial Basic Research Program (Natural Science) (grant number QKHJCZK [2022] YB305), and the Guizhou Normal University Academic Emerging Talent Fund Program (Grant number QSXM [2022] 14).
Author information
Authors and Affiliations
Contributions
LG and XYD conceived research plans and designed experiments. LG, XXC, GYW, YYS and YYH conducted experiments. LG, YYH and YYS wrote the draft. HCW and BZ analyzed the data. LG, YYH and XYD reviewed and edited this article. The authors declare no conflicts of interest.
Corresponding author
Ethics declarations
Conflict of interest
We declare that we have no conflicts of interest.
Additional information
Communicated by Ting-Ting Yuan.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gu, L., Hou, Y., Sun, Y. et al. The maize WRKY transcription factor ZmWRKY64 confers cadmium tolerance in Arabidopsis and maize (Zea mays L.). Plant Cell Rep 43, 44 (2024). https://doi.org/10.1007/s00299-023-03112-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00299-023-03112-8