Abstract
Key message
Rice-produced SD1 retains its physicochemical properties and provides efficient pre-exposure HIV-1 prophylaxis against infection in vitro.
Abstract
Scytovirin (SVN) is an HIV-neutralizing lectin that features two structural domains (SD1 and SD2) that bind to HIV-1 envelope glycoproteins. We expressed SD1 in rice seeds as a potential large-scale production platform and confirmed that rice-derived SD1 binds the HIV-1 envelope glycoprotein gp120 in vitro. We analyzed the thermodynamic properties of SD1 compared to full-size SVN (produced in E. coli) by isothermal titration and differential scanning calorimetry to characterize the specific interactions between SVN/SD1 and gp120 as well as to high-mannose oligosaccharides. SVN bound with moderate affinity (Kd = 1.5 µM) to recombinant gp120, with 2.5-fold weaker affinity to nonamannoside (Kd of 3.9 µM), and with tenfold weaker affinity to tetramannoside (13.8 µM). The melting temperature (Tm) of full-size SVN was 59.1 °C and the enthalpy of unfolding (ΔHunf) was 16.4 kcal/mol, but the Tm fell when SVN bound to nonamannoside (56.5 °C) and twice as much energy was required for unfolding (ΔHunf = 33.5 kcal/mol). Interestingly, binding to tetramannoside destabilized the structure of SD1 (ΔTm ~ 11.5 °C) and doubled the enthalpy of unfolding, suggesting a dimerization event. The similar melting phenomenon shared by SVN and SD1 in the presence of oligomannose confirmed their conserved oligosaccharide-binding mechanisms. SD1 expressed in transgenic rice was able to neutralize HIV-1 in vitro. SD1 expressed in rice, therefore, is suitable as a microbicide component.
Similar content being viewed by others
Introduction
Lectins are carbohydrate-binding proteins that bind reversibly to specific glycan structures (Mitchell et al. 2017; Singh et al. 2017). Many lectins recognize the N-linked glycans present on viral glycoproteins (François and Balzarini 2012; Kumar et al. 2012) and can, therefore, prevent viral proteins from interacting with receptors on the target cell surface, blocking the uptake of viral particles and interrupting the infection cycle (Balzarini 2006; Botos and Wlodawer 2005).
A recognized target of antiviral lectins is the human immunodeficiency virus (HIV), which has thus far infected ~ 76 million people, ~ 38 million of whom are still living with the virus today (WHO 2020). HIV-1 is the most common and virulent type of HIV. It enters susceptible cells when the viral surface glycoprotein gp120 interacts with the receptor CD4 on the surface of lymphocytes. This is followed by the binding of co-receptors (CCR5 or CXCR4), allowing the transmembrane glycoprotein gp41 to induce membrane fusion (Steckbeck et al. 2013). Molecules such as lectins, that bind gp120 and/or gp41, can, therefore, act as HIV-1 entry inhibitors and may be suitable as topical microbicides, representing a subset of pre-exposure prophylaxis strategies (Xion et al. 2006; Vamvaka et al. 2018). Lectins with the potential to act as HIV-1 entry inhibitors include cyanovirin-N (CV-N) from the cyanobacterium Nostoc ellipsosporum (Boyd et al. 1997) and griffithsin (GRFT) from the red alga Griffithsia sp. (Mori et al. 2005), both of which bind oligomannose structures on gp120 and can neutralize the virus before target cell uptake (Mori et al. 2002). Both lectins have been investigated as topical microbicides to prevent HIV-1 transmission (Mori et al. 2005; O’Keefe et al. 2009; Moulaei et al. 2010; Vamvaka et al. 2016b, 2016c) and GRFT has recently entered human clinical trials (ClinicalTrials.gov identifier: NCT02875119 and NCT04032717). Another microbicide candidate is scytovirin (SVN) from the cyanobacterium Scytonema varium (Bokesch et al. 2003). The 95-amino-acid SVN polypeptide has a molecular weight of 9.7 kDa and contains two structural domains, namely SD1 (residues 1–48) and SD2 (residues 49–95), linked by five intra-chain disulfide bonds (McFeeters et al. 2007; Moulaei et al. 2007). SVN can neutralize HIV-1 by binding to gp120, gp160 and gp41. SD1 has demonstrated similar activity to full-length SVN, whereas SD2 is less potent than the full-length SVN and SD1 (Xiong et al. 2006). The domains have different affinities for HIV-1 and bind to carbohydrate ligands independently. Native SVN is active against both laboratory strains and primary isolates of HIV-1, the latter providing a more faithful representation of the inoculum that humans encounter during HIV-1 transmission, with EC50 values ranging from 0.3 to 22 nM (Bokesch et al. 2003).
To help prevent the spread of HIV, viral entry inhibitors such as broadly neutralizing monoclonal antibodies and lectins have been formulated as microbicides for pre-exposure prophylaxis, and have been tested in vitro, in animal studies, and in human clinical trials (O’Keefe et al. 2010; Ramessar et al. 2010; McCoy and Weiss 2013; Bar et al. 2016). Entry inhibitors can be produced as recombinant proteins by fermentation in mammalian cells (antibodies) or microbial systems (lectins), but these platforms are too expensive for the large-scale and low-margin production campaigns needed to manufacture microbicides for target populations that are disproportionately found in countries with small budgets for public health (Ma et al. 2003; Stoger et al. 2005; Mir‐Artigues et al. 2019). The demand is created by the large size of the at-risk population, the frequency of microbicide application, and the large doses required for efficacy (Ramessar et al. 2008; Ma et al. 2013; Sabalza et al. 2014). Transgenic plants can address these drawbacks, allowing inexpensive microbicide production (Capell et al. 2020; Lobato Gomez et al. 2021; He et al. 2021). In the context of microbicides, transgenic cereals offer additional advantages because lectins and/or antibodies can be produced locally and stored/transported as dry seed (obviating the need for a cold chain), and the products can be administered directly as crude extracts prior to intercourse, thus eliminating the costs of downstream processing (Ramessar et al. 2010; Stoger et al. 2005; Sabalza et al. 2013). However, before transgenic plants can be used for the large-scale production of microbicidal lectins, it is necessary to ensure the recombinant proteins remain functional by characterizing their physicochemical interactions.
Here we investigated the suitability of the lectin SD1 as a plant-derived microbicide component. Using isothermal titration calorimetry (ITC), that measures the heat of association for binding interactions between two macromolecules, and by differential scanning calorimetry (DSC), which measures the heat change associated with the thermal denaturation of a molecule when heated at a constant rate, we compared the thermodynamic behavior of SD1 to full-size SVN produced in E. coli. We analyzed the binding of SD1 and SVN to HIV-1 envelope glycoprotein gp120 as well as individual high-mannose oligosaccharides. The crude plant extract containing SD1 was also tested for HIV-binding activity and its ability to prevent HIV-1 infection in vitro using the TRO11 pseudovirus, which covers the diversity of most circulating strains of HIV-1 group M subtype B, as well as primary isolates of HIV-1.
Materials and methods
Genetic constructs and transgenic plants
The S. varium SD1 gene in vector pET-28a ( +) (National Cancer Institute, Frederick, MD, USA) was amplified by PCR in a 25 µL reaction containing 0.125 µL GoTaq G2 Flexi DNA polymerase (5 U/µL) in 1 × Green GoTaq Reaction Buffer (Promega, Madison, WI, USA), 0.5 µL (10 µM) forward primer 5′-GGA TCC ATG AGC GAT AAA ATT ATT C-3′ (BamHI restriction site underlined) and reverse primer 5′-GAA TTC GCA GCC GGA TCT CAG TGG TG-3′ (EcoRI restriction site underlined). The reaction was heated to 95 °C for 2 min followed by 35 cycles of 95 °C for 45 s, 60 °C for 45 s and 72 °C for 5 min and a final extension step at 72 °C for 10 min. The product was transferred to the shuttle vector pGEM-T easy (Promega) and introduced into competent Escherichia coli cells, which were incubated overnight at 37 °C with ampicillin for selection. Plasmid DNA from positive colonies was verified by sequencing (StabVida, Caparica, Portugal). Plasmid DNA was digested with BamHI and EcoRI (Promega) and the SD1 gene was transferred to vector pRP5 containing the endosperm-specific rice prolamin promoter (Naqvi et al. 2009) and the nos terminator.
Generation of transgenic rice plants and confirmation of SD1 gene expression
Seven-day-old mature rice zygotic embryos (Oryza sativa cv. Nipponbare) were transferred to Murashige and Skoog (MS) osmotic medium (4.4 g/L MS salts supplemented with 0.3 g/L casein hydrolysate, 0.5 g/L proline, 72.8 g/L mannitol and 30 g/L sucrose) 4 h before bombardment with 10 mg gold particles coated with the construct carrying the SD1 transgene and second construct carrying the selectable marker hpt. The embryos were returned to MS osmotic medium for 12 h before selection on standard MS medium (4.4 g/L MS salts, 0.3 g/L casein, 0.5 g/L proline and 30 g/L sucrose) supplemented with 50 mg/L hygromycin and 2.5 mg/L 2,4-dichlorophenoxyacetic acid in the dark for 2–3 weeks. Transgenic plantlets were regenerated and hardened off in soil. Plants were grown in the greenhouse or growth chamber at 28/20 °C day/night temperature with a 10 h photoperiod and 60–90% relative humidity for the first 50 days, followed by maintenance at 21/18 °C day/night temperature with a 16 h photoperiod thereafter in a growth chamber.
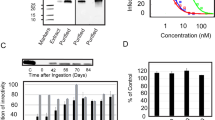
To confirm and quantify SD1 transgene expression in transgenic plants, total RNA was extracted from 21-day-old immature seeds using the LiCl method and cDNA was prepared using the QuantiTect reverse transcription kit (Qiagen, USA). Each 25 μl reaction contained 10 ng cDNA, 1 × SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA), 0.2 μM forward primer (5′-ACG GAT GTA CTC AAA GCG GAC-3′) and 0.2 μM reverse primer (5′-TAT GGC ATC CGT GGT ATC CCG-3′). Quantitative PCR was carried out by heating the reaction to 95 °C for 3 min followed by 39 cycles of 95 °C for 10 s, 59 °C for 30 s and 72 °C for 20 s and a final step at 95 °C for 10 s using CFX96 system (Bio-Rad Laboratories). Amplification specificity was confirmed by melt curve analysis on the final PCR products in the temperature range 50–90 °C with fluorescence acquired after each 0.5 °C increment. Values represent the mean of three technical replicates. The relative expression was calculated using the 2-ΔCt formula, where ΔCt is the threshold cycle difference between the actin and SD1 genes.
Quantification of SD1 protein and in vitro analysis of binding to gp120
The presence of SD1 in transgenic rice endosperm was confirmed by enzyme-linked immunosorbent assay (ELISA). Mature rice seeds (25 seeds per sample, equivalent to 375 mg) were ground in three volumes of phosphate-buffered saline (PBS, pH 7.4) and centrifuged twice (13,000×g, 10 min, 4 °C). ELISA plates were coated with 50 ng/well of recombinant gp120 from HIV-1 strain IIIB provided by the MRC Centralized Facility for AIDS Reagents (Potters Bar, UK) overnight at 4 °C. The plates were then washed with PBS and incubated for 2 h at room temperature with blocking solution comprising 1% bovine serum albumin (BSA; Sigma–Aldrich, Madrid, Spain) in PBS containing 0.1% Tween-20 (PBST). SD1 was detected in serial dilutions of seed extract using a primary rabbit anti-SVN polyclonal antiserum (National Cancer Institute, Bethesda, MD, USA) diluted 1:2000 and a secondary horseradish peroxidase (HRP)-conjugated anti-rabbit IgG antibody (Sigma–Aldrich) diluted 1:10,000. HRP activity was detected by staining with 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Thermo Fisher Scientific, Waltham, MA, USA). The reaction was stopped with 0.16 M H2SO4, and absorbance was measured at 450 nm. Recombinant SD1 purified from E. coli (Xion et al. 2006) was used as a positive control and wild-type rice extract and 1% BSA in PBS as negative controls. The yield of SD1 in rice seeds (ng/g dry seed) was calculated as the product of the OD450 value, taking the rice extract dilution factors into consideration and the volume of buffer used for the protein extraction, divided by the weight of the seeds. Known concentrations of SD1 purified from E. coli were used to perform the standard curve. The specific antigen-binding activity of SD1 was determined using the same procedure.
Isothermal titration calorimetry
ITC was carried out using a VP-ITC device (Malvern Panalytical, Malvern, UK). For the SVN:gp120 titration experiments, 102 µM E. coli-produced SVN (Xion et al. 2006) was titrated into a calorimetry cell containing 1.6 µM gp120. For the sugar titration experiments, 2.4 mM nonamannoside or 1 mM tetramannoside was titrated into a calorimetry cell containing 102 µM SVN. Purified oligosaccharides were either purchased from Glyko Inc. (Novato, CA) or were the kind gift of Dr. Peter Seeberger (Max Planck Institute) and were synthesized as previously described (Ratner et al. 2002). In a typical experiment, 5 µL aliquots of titrant were injected into a rapidly mixing (300 rpm) solution in the calorimetry cell (volume = 1.4426 mL) with a total of 55 injections during the experiment. Controls were prepared with identical amounts of titrant injected into a protein-free buffer, and control values were subtracted from the results of the other experiments. Titrations were carried out at 30 °C in 10 mM sodium phosphate buffer (pH 7.4) containing 3 mM sodium azide. The isotherm, corrected for dilution/buffer effects, was fitted to a nonlinear least squares curve-fitting model (for a 1-set of identical sites) using Microcal Origin v7.0 (OriginLab, Northampton, MA, USA). We extracted values for enthalpy, binding affinity and stoichiometry from the binding curve, and the free energy and entropy of interaction were calculated using Eqs. (1) and (2):
where ∆G is the change in Gibbs free energy, R is the gas constant (~ 1.987 cal/mol K), T is the absolute temperature (303 K), Ka is the equilibrium constant, ∆H is the change in enthalpy and ∆S is the change in entropy.
Differential scanning calorimetry
DSC was carried out using a VP-DSC device (Malvern Instruments). In a typical experiment, the concentration of E. coli-produced SVN or SD1 (Xion et al. 2006) was adjusted to 20 µM and was prepared in a total volume of 1.0 mL with or without sugars in 10 mM sodium phosphate buffer (pH 7.4) containing 3 mM sodium azide. Before adding the samples, the degassed buffer solutions were first placed in the sample and reference cells. Following the first up scan (10–95 °C) and during the first down scan (at 25 °C), the sample cell was emptied and filled with a degassed SVN (or SD1) solution. The sample and cell compartments were capped with adjusted positive pressure (30 psi) and allowed to progress through eight alternating up and down scans at 60 °C/h with a filter period of 16 s. The completed sample thermogram was corrected for buffer effects (subtracting the first up scan) followed by normalized correction of the absolute baseline and cubic approximation of the pre-transition (structured) and post-transition (unstructured) regions of the thermogram. The baseline-corrected thermogram was fitted to a non-2-state unfolding model and the fitting error was evaluated for single versus multiple unfolding units. From this model-fitting, values for melting temperature (Tm) and enthalpy of unfolding (∆Hcal) were extrapolated for each transition. The reversibility of melting was assessed by superimposing the repeated up-scans. For compound experiments, the post-titrated solutions of SVN (or SD1) with sugars were adjusted to a final protein concentration of 20 µM. Similar parameters and procedures were used to assess changes to Tm and the enthalpy of unfolding.
HIV-neutralization assays
HIV-1 pseudoviruses BaL (laboratory isolate), TRO11 and AC10 (primary isolates) were generated by the co-transfection of HEK293T cells with HIV-1 Env protein-expressing plasmids (NIH AIDS Reagent Program, Manassas, VA, USA) and the PSG3 vector as previously described (Sánchez-Palomino et al. 2011). The supernatants were harvested 24 h post-transfection and were passed through a 0.45 μm filter and stored at − 80 °C. Viral titration and neutralization were tested using a standard TZM-bl assay. Briefly, serially diluted samples were pre-incubated in duplicate with 200 TCID50 of pseudovirus stock for 1 h at 37 °C in 96-well culture plates. We then added 10,000 TZM-bl luciferase-reporter cells to each well. The plates were incubated at 37 °C in a 5% CO2 atmosphere for 48 h in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) and 30 μg/ml dextran (Sigma–Aldrich). The luciferase substrate Britelite Plus (PerkinElmer, Waltham, MA, USA) was used for the readout. Percent neutralization was determined by calculating the difference in average relative light units (RLUs) between virus control (cells + virus) and test wells (cells + sample + virus). Neutralizing titers are expressed as the concentration of total protein or SD1 necessary to reduce the RLUs by 50% (IC50). All data were fitted to an inhibitor vs response (three parameters) model using GraphPad Prism v6 (GraphPad Software, San Diego, CA, USA).
Results
Expression of SD1 in transgenic plants and gp120-binding activity in vitro
Mature seed-derived rice embryos were co-transformed with a construct containing the SD1 coding sequence under the control of an endosperm-specific promoter, plus a second construct containing the selectable marker hpt (Christou et al. 1991). Embryo-derived callus was selected on hygromycin-supplemented medium and independent transformants were regenerated and transferred to the greenhouse. Total RNA extracted from T1 seeds was reverse transcribed using SD1-specific primers and amplified by qPCR to confirm transgene expression. The presence of correctly assembled SD1 protein in the endosperm was confirmed by ELISA and the yield was determined by calculating the concentrations from titration curves based on positive controls spiked with known amounts of SD1 produced in E. coli and non-transformed rice endosperm as a negative control. We selected the line which accumulated SD1 at the highest level (26 ng/g dry seed weight) for subsequent experiments. Crude endosperm extracts from this transgenic line were tested for their ability to bind HIV gp120IIIB in vitro, using SD1 produced in E. coli as positive control and wild-type rice endosperm extracts as a negative control. We observed similar concentration-dependent gp120 binding in the positive control and in the extracts of the transgenic seeds (Fig. 1).
Rice endosperm extracts (containing SD1) bind to gp120. Plates were coated with gp120 (50 ng/well). SD1 was detected with a primary rabbit anti-SVN polyclonal antiserum. Lead Transgenic Line: starting concentration corresponds to 0.5 dilution of the original endosperm extract. WT: starting concentration corresponds to 0.5 dilution of the original endosperm extract. SD1: purified from E. coli and used as a positive control (starting concentration was 4 ng/mL). IB: 1% BSA in PBST. A fourfold dilution series were performed. Data are means ± standard errors (n = 3). Turkey test (ANOVA) with a P ≤ 0.05 was performed. Asterisks (*) indicate significant differences and ns indicates no significant differences comparing to each dilution of the Lead Transgenic Line
Thermodynamic analysis of E. coli-produced SVN and SD1 binding interactions
The ITC isotherms (Fig. 2) were fit to a nonlinear least squares curve-fitting model for identical binding sites. The values for enthalpy, binding affinity were extracted directly from this curve fitted model (shown in red, Fig. 2) and the free energy and entropy of interaction were back calculated using Eqs. 1, 2 (“Materials and Methods”). All thermodynamic parameters extracted from this curve-fitting analysis are listed in Table 1. The mid-point of the transitions (Fig. 2) extrapolated to the X-axis indicates the stoichiometry of the interaction between SVN and its binding partners. As can be seen in Table 1, the binding between SVN and gp120 was driven by both favorable enthalpic and entropic contributions (negative ΔH value and positive ΔS value, respectively) and resulted in a moderate binding affinity of Kd = 1.63 µM. The stoichiometry of the interaction was 5:1, five molecules of SVN binding to a single molecule of gp120 (Fig. 2A). In contrast, SVN bound to oligomannosides with 1:1 stoichiometry (Fig. 2B and C) and the binding was predominantly driven entropically (positive ΔS value; Table 1). As expected, the enthalpy of binding was significantly lower when SVN bound to the oligosaccharides rather than gp120, presumably because fewer surface contacts are available on the smaller molecules. Accordingly, the enthalpy–entropy compensation reduced the free energies of binding (ΔG), resulting in a ~ 2.5-fold lower affinity for nonamannoside (Kd = 4.0 µM) and a ~ tenfold lower affinity for tetramannoside (Kd = 17.8 µM) (Table 1).
DSC results showed SVN melted at Tm = 59.1 °C and had an enthalpy of unfolding (ΔH) of 16.4 kcal/mol (Table 2). In the presence of nonamannoside, the oligosaccharide–protein complex melted at a lower temperature (Tm = 56.5 °C) but required twice as much energy (ΔH = 33.5 kcal/mol) for unfolding (Fig. 2B and C). Similarly, DSC experiments with SD1 showed that binding to tetramannoside destabilized the SD1 structure and caused a twofold increase in the enthalpy of unfolding (Fig. 3). These data suggest that the tertiary structures of SVN and SD1 were destabilized (lower Tm) when binding the oligosaccharides, favoring the dimerization of the oligosaccharide–protein complexes. The ability of SVN and SD1 to oligomerize via crosslinking with sugars is consistent with other anti-HIV lectins such as CV-N (Shenoy et al. 2002).
Neutralization of HIV pseudoviruses in vitro by crude rice endosperm extracts containing SD1
The activity of extracts from the transgenic line was tested in HIV infectivity and neutralization assays with three HIV pseudoviruses expressing the envelope glycoprotein of the laboratory adapted isolate BaL or the primary isolates TRO11 and AC10. Wild-type rice endosperm extracts were used as a negative control. The extract of the transgenic seeds was able to neutralize all three pseudoviruses with IC50 values ranging from 0.10 to 0.16 mg protein/mL, corresponding to apparent IC50 values for SD1 ranging from 0.29 to 0.45 ng/mL (Fig. 4). Interestingly, the wild-type rice extract also demonstrated significant neutralization activity at the highest concentration tested, with IC50 values ranging from 0.32 to 0.38 mg protein/mL, indicating that components of the rice endosperm can reduce virus infectivity independently of the lectin (Fig. 4).
Neutralization activity of the extract from Lead Transgenic Line expressing SD1 (SD1) and wild-type rice extract (Ctrol) in a pseudovirus assay using BaL, TRO11 and AC10 isolates. Extracts were normalized by the total amount of protein (x-axis). The calculated concentration of SD1 is also shown in the additional red x-axis. Apparent SD1 IC50 values are indicated for each pseudovirus. Data are means ± standard errors (n = 2). The experiment was carried out twice with the same results. Notice that some error bars are too small to be visible at this scale
Discussion
The biological function of many lectins is associated with self-recognition and cell adhesion in multicellular organisms. Lectins from natural sources have been shown to bind to the N-linked glycans on the surface of many enveloped viruses (Goldstein and Poretz 1986; François and Balzarini 2012; Kumar et al. 2012) resulting in the disruption of interactions between proteins on the viral envelope and their cellular receptors (Balzarini 2006; Botos and Wlodawer 2005; Sacchettini et al. 2001; Shenoy et al. 2002). This characteristic makes them useful as broad-spectrum antiviral agents, particularly for use in pre-exposure prophylaxis (O’Keefe et al. 2010; Ramessar et al. 2010; McCoy and Weiss 2013; Bar et al. 2016). For example, SVN from the cyanobacterium S. varium (Bokesch et al. 2003) binds to mannose-rich oligosaccharides on the envelope glycoproteins of several viruses, including Ebola (Garrison et al. 2014), dengue and other flaviviruses with the same glycoprotein pattern (Siqueira et al. 2017), hepatitis C virus (Takebe et al. 2013), and HIV-1 (Xiong et al. 2006).
SVN has been proposed as a candidate HIV entry inhibitor for therapeutic and prophylactic applications (McFeeters et al. 2007). It has been expressed for this purpose in E. coli (Xiong et al. 2006) and Lactobacillus plantarum (Janahi et al. 2018), with both bacterial recombinant proteins able to bind HIV-1 gp160 but the version produced in E. coli demonstrating marginally higher potency against HIV-1 in a TZM-bl cell assay: ~ 89% reduction in infectivity after 48 h compared to ~ 86% for the version produced in L. plantarum (Janahi et al. 2018). SVN is particularly active against HIV because it can bind to multiple sites on gp120 (Bokesch et al. 2003; Siqueira et al. 2017). Specifically, SVN binds to α1,2-α1,2-α1,6-linked tetramannoside (Adams et al. 2004) as well larger oligosaccharides such as Man-8 and Man-9 on gp120 and gp41 (Xiong et al. 2006). The ability of SVN to bind multiple sites reflects the presence of two domains (SD1 and SD2), with partially conserved tertiary structures, which can each bind independently to oligosaccharides. The combination of multisite and multivalent binding by SVN increases its potency. When these domains were expressed separately in E. coli, SD1 showed equivalent antiviral activity to full-size SVN whereas SD2 bound to gp120 with 50% lower affinity (Xiong et al. 2006), which may reflect the flipped orientation of the middle-turn regions within the individual carbohydrate-binding domains (McFeeters et al. 2007).
Transgenic plants offer an inexpensive and scalable production platform for lectins as a step toward the development of microbicidal cocktails based on plant extracts (Ma et al. 2003; Stoger et al. 2005; Ramessar et al. 2008; Lobato Gomez et al. 2021). The seeds of cereals such as rice are particularly suitable because they have “generally regarded as safe” status, allowing the direct application of extracts to the mucosal surface, and recombinant proteins in dry seeds remain stable for many years, thus addressing the limited availability of cold chains in developing countries (Daniell et al. 2001; Ramessar et al. 2008; Ma et al. 2013; Arcalis et al. 2014). We have previously used rice to express the HIV-neutralizing antibody 2G12 (Vamvaka et al. 2016a) and the lectins GRFT (Vamvaka et al. 2016b) and CV-N (Vamvaka et al. 2016c), as well as all three of these components simultaneously (Vamvaka et al. 2018). We found that the recombinant proteins retained their physicochemical properties and biological activity when compared to positive controls produced in E. coli. Before using transgenic plants for the large-scale production of SVN and/or its smaller congener SD1, it was necessary to conduct similar tests to ensure that the recombinant proteins can still bind with high affinity to HIV glycoproteins and neutralize the virus in vitro.
We compared the biological activity of recombinant SD1 proteins produced in E. coli and rice by testing their ability to bind gp120 in vitro and found that both versions showed comparable binding activity. Rice-produced SD1 showed the same concentration-depending affinity for gp120 as E. coli SD1 (the starting concentration of SD1 protein tested by ELISA (Fig. 1) was 4 ng/mL in both cases) indicating that the rice-produced protein has the same oligosaccharide-dependent binding properties as the bacterial-expressed protein (Xiong et al. 2006). We probed the physicochemical basis of this binding activity in ITC and DSC experiments to gain more insight into the specific interactions between E. coli-produced SVN/SD1 and HIV envelope glycoproteins and/or high-mannose oligosaccharides. ITC revealed a Kd value of 1.5 µM for SVN binding to gp120, and the post-titrated solution was clear, ruling out the presence of insoluble protein–protein aggregates. The binding of SVN to gp120 resulted in a 5:1 stoichiometry, which was the same as previously reported for CV-N (O’Keefe et al. 2000). This suggests that both lectins may recognize similar or identical surface contacts on gp120. Previous studies using microarrays indicated that SVN binds terminal α1,2-mannose on the D3 arm of gp120 and can, therefore, recognize Man-9, Man-8 and even a tetramannoside structure mimicking the D3 arms of Man-9 (Ratner and Seeberger 2007), but not Man-7, which lacks a terminal α1,2-mannose. Accordingly, we characterized the binding of SVN to tetramannoside and nonamannoside (similar to Man-9 but missing the core GlcNAc residues) by ITC. SVN bound to both structures with 1:1 stoichiometry but with a significant difference in affinity between the nonamannoside (Kd = 4 µM) and tetramannoside (Kd = 18 µM). SVN binding to nonamannoside was enthalpically more favorable (ΔH = –5.3 kcal/mol) than tetramannoside (ΔH = –1.9 kcal/mol), suggesting the presence of more polar/electrostatic, van der Waals and hydrogen bond contacts on nonamannoside compared to the smaller sugar structure. In addition, nonamannoside is a branched sugar and as such can engage in multivalent interactions with SVN via its multiple α1,2-mannose termini, this may account for the stronger binding compared to tetramannoside. Interestingly, a baseline disturbance was noted in the nonamannoside-SVN ITC experiment, suggesting that soluble high-molecular-weight complexes may have formed during the titration. It is also likely that SD1 and SD2 engage in multisite binding, which combined with the multivalency of nonamannoside can explain the higher affinity for this oligosaccharide while maintaining an overall 1:1 stoichiometry. However, if such higher-order structures were formed, they did not persist, hence the absence of aggregation or cloudiness in the post-titrated solution. The difference in carbohydrate-binding strength between SD1/SD2 and the domains of CV-N may be the reason SVN cannot lock in the higher-order cross-linked structures with branched oligosaccharides. This may be the real reason for its inability to achieve the type of potent irreversible binding and inactivation of gp120 that has been observed for CV-N (Xiong et al. 2006).
DSC experiments indicated that SVN melted at a moderate temperature (Tm = 59.1 °C) and that the enthalpy of unfolding (ΔH) was 16.4 kcal/mol, as would be expected for a 10-kDa protein. In the presence of nonamannoside, the oligosaccharide–protein complex melted at a lower temperature (Tm = 56.5 °C), but required twice as much energy (ΔH = 33.5 kcal/mol) for unfolding. The lower melting temperature suggests that at least some part of the tertiary structure of SVN was destabilized in the presence of the sugar and that either the destabilized structure had exposed hydrophobic surface patches that allowed for potential protein–protein dimerization contacts, or that the sugar itself facilitated crosslinking during dimerization. A similar phenomenon was observed in the DSC experiments with SD1, where binding to tetramannoside destabilized the SD1 structure (ΔTm ~ 11.5 °C) and resulted in a twofold increase in the enthalpy of unfolding. Additional structural studies may resolve the nature of the destabilization mediated by the oligosaccharide binding and the resulting dimerization event. Such studies may also help to determine whether the dimeric structure is entirely composed of protein or is bridged by an oligosaccharide. Our DSC results show that the full-length SVN and its congener SD1 both melt in a similar manner upon binding to oligomannose sugars, suggesting that they may share an analogous mode of interaction with the sugars on gp120 and may, therefore, demonstrate similar HIV-neutralizing activities.
Accordingly, we tested the neutralization activity of rice-expressed SD1 in an HIV-1 pseudovirus assay based on three different Env glycoprotein variants, a laboratory adapted strain and two primary isolates representing the diversity of most circulating HIV-1 strains of group M subtype B (Hemelaar et al. 2006). SD1 from rice exhibited nanomolar activity against HIV and protected cells from infection (IC50 = 0.29 to 0.45 ng/mL). Interestingly, the wild-type crude extract also showed modest HIV-neutralizing activity even though there was no significant binding to HIV-1 gp120, with an IC50 2.4–3.8 times higher than the one obtained for the extract of the transgenic seeds (Fig. 1), suggesting that virus neutralization may be an indirect effect perhaps involving interactions with the host cell surface or molecular crowding (Vamvaka et al. 2018).
In conclusion, our data suggest that rice-derived SD1 retains its physicochemical and biological properties and is, therefore, a promising candidate microbicide for pre-exposure HIV-1 prophylaxis. SD1 has fewer disulfide bonds than SVN and is smaller and, therefore, probably less immunogenic. Its low nanomolar activity and physical stability are major advantages for a microbicide component (Xiong et al. 2006). The functionality of SD1 in the crude rice extract and the minimal processing and purification steps required during manufacturing will reduce production costs significantly compared to traditional cell-based platforms.
References
Adams EW, Ratner DM, Bokesch HR, McMahon JB, O’Keefe BR, Seeberger PH (2004) Oligosaccharide and glycoprotein microarrays as tools in HIV glycobiology: glycan-dependent gp120/protein interactions. Chem Biol 11:875–881. https://doi.org/10.1016/j.chembiol.2004.04.010
Arcalis E, Ibl V, Peters J, Melnik S, Stoger E (2014) The dynamic behavior of storage organelles in developing cereal seeds and its impact on the production of recombinant proteins. Front Plant Sci 5:439. https://doi.org/10.3389/fpls.2014.00439
Balzarini J (2006) Inhibition of HIV entry by carbohydrate-binding proteins. Antiviral Res 71:237–247. https://doi.org/10.1016/j.antiviral.2006.02.004
Bar KJ, Sneller MC, Harrison LJ, Justement JS, Overton ET, Petrone ME, Salantes DB, Seamon CA, Scheinfeld B, Kwan RW (2016) Effect of HIV antibody VRC01 on viral rebound after treatment interruption. N Engl J Med 375:2037–2050. https://doi.org/10.1056/NEJMoa1608243
Bokesch HR, O’Keefe BR, McKee TC, Pannell LK, Patterson GML, Gardella RS, Sowder RC, Turpin J, Watson K, Buckheit RW (2003) A potent novel anti-HIV protein from the cultured cyanobacterium Scytonema varium. Biochemistry 42:2578–2584. https://doi.org/10.1021/bi0205698
Botos I, Wlodawer A (2005) Proteins that bind high-mannose sugars of the HIV envelope. Prog Biophys Mol Biol 88:233–282. https://doi.org/10.1016/j.pbiomolbio.2004.05.001
Boyd MR, Gustafson KR, McMahon JB, Shoemaker RH, O’Keefe BR, Mori T, Gulakowski RJ, Wu L, Rivera MI, Laurencot CM (1997) Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: potential applications to microbicide development. Antimicrob Agents Chemother 41:1521–1530. https://doi.org/10.1128/AAC.41.7.1521
Capell T, Twyman RM, Armario-Najera V, Ma JK-C, Schillberg S, Christou P (2020) Potential applications of plant biotechnology against SARS-CoV-2. Trends Plant Sci 25:635–643. https://doi.org/10.1016/j.tplants.2020.04.009
Christou P, Ford TL, Kofron M (1991) Production of transgenic rice (Oryza sativa L.) plants from agronomically important indica and japonica varieties via electric discharge particle acceleration of exogenous DNA into immature zygotic embryos. Bio/technology 9:957–962. https://doi.org/10.1038/nbt1091-957
Daniell H, Streatfield SJ, Wycoff K (2001) Medical molecular farming: production of antibodies, biopharmaceuticals and edible vaccines in plants. Trends Plant Sci 6:219–226. https://doi.org/10.1016/s1360-1385(01)01922-7
François KO, Balzarini J (2012) Potential of carbohydrate-binding agents as therapeutics against enveloped viruses. Med Res Rev 32:349–387. https://doi.org/10.1002/med.20216
Garrison AR, Giomarelli BG, Lear-Rooney CM, Saucedo CJ, Yellayi S, Krumpe LRH, Rose M, Paragas J, Bray M, Olinger GG Jr (2014) The cyanobacterial lectin scytovirin displays potent in vitro and in vivo activity against Zaire Ebola virus. Antiviral Res 112:1–7. https://doi.org/10.1016/j.antiviral.2014.09.012
Goldstein IJ, Poretz RD (1986) Isolation and chemical properties of lectins. In: Liener IE, Sharon N, Goldstein IT (eds) The lectins: properties, functions and applications in biology and medicine. Academic Press, New York, pp 33–247. https://doi.org/10.1016/B978-0-12-449945-4.50007-5
He W, Baysal C, Lobato Gómez M, Huang X, Alvarez D, Zhu C, Armario-Najera V, Blanco Perera A, Cerda Bennaser P, Saba-Mayoral A, Sobrino-Mengual G, Vargheese A, Abranches R, Alexandra Abreu I, Balamurugan S, Bock R, Buyel JF, da Cunha NB, Daniell H, Faller R, Folgado A, Gowtham I, Häkkinen ST, Kumar S, Sathish Kumar R, Lacorte C, Lomonossoff GP, Luís IM, Ma K-CJ, McDonald KA, Murad A, Nandi S, O’Keef B, Parthiban S, Paul MJ, Ponndorf D, Rech E, Rodrigues JCM, Ruf S, Schillberg S, Schwestka J, Shah PS, Singh R, Stoger E, Twyman RM, Varghese IP, Vianna GR, Webster G, Wilbers RHP, Christou P, Oksman-Caldentey KM, Capell T (2021) Contributions of the international plant science community to the fight against infectious diseases in humans-part 2: affordable drugs in edible plants for endemic and re-emerging diseases. Plant Biotechnol J 19(10):1921–1936. https://doi.org/10.1111/pbi.13658
Hemelaar J, Gouws E, Ghys PD, Osmanov S (2006) Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS 20:13–23. https://doi.org/10.1097/01.aids.0000247564.73009.bc
Janahi EMA, Haque S, Akhter N, Wahid M, Jawed A, Mandal RK, Lohani M, Areeshi MY, Almalki S, Das S (2018) Bioengineered intravaginal isolate of Lactobacillus plantarum expresses algal lectin scytovirin demonstrating anti-HIV-1 activity. Microb Pathog 122:1–6. https://doi.org/10.1016/j.micpath.2018.06.002
Kumar KK, Chandra KLP, Sumanthi J, Reddy GS, Shekar PC, Reddy BVR (2012) Biological role of lectins: a review. J Orofac Sci 4:20. https://doi.org/10.4103/0975-8844.99883
Lobato Gómez M, Huang X, Alvarez D, He W, Baysal C, Zhu C, Armario-Najera V, Blanco Perera A, Cerda Bennasser P, Saba-Mayoral A, Sobrino-Mengual G, Vargheese A, Abranches R, Abreu IA, Balamurugan S, Bock R, Buyel JF, da Cunha NB, Daniell H, Faller R, Folgado A, Gowtham I, Häkkinen ST, Kumar S, Ramalingam SK, Lacorte C, Lomonossoff GP, Luís IM, Ma JK, McDonald KA, Murad A, Nandi S, O’Keefe B, Oksman-Caldentey KM, Parthiban S, Paul MJ, Ponndorf D, Rech E, Rodrigues JCM, Ruf S, Schillberg S, Schwestka J, Shah PS, Singh R, Stoger E, Twyman RM, Varghese IP, Vianna GR, Webster G, Wilbers RHP, Capell T, Christou P (2021) Contributions of the international plant science community to the fight against human infectious diseases—part 1: epidemic and pandemic diseases. Plant Biotechnol J 19(10):1901–1920. https://doi.org/10.1111/pbi.13657
Ma JK-C, Drake PMW, Christou P (2003) The production of recombinant pharmaceutical proteins in plants. Nat Rev Genet 4:794–805. https://doi.org/10.1038/nrg1177
Ma JK, Christou P, Chikwamba R, Haydon H, Paul M, Ferrer MP, Ramalingam S, Rech E, Rybicki E, Wigdorowitz A (2013) Realising the value of plant molecular pharming to benefit the poor in developing countries and emerging economies. Plant Biotechnol J 11:1029–1033. https://doi.org/10.1111/pbi.12127
McCoy LE, Weiss RA (2013) Neutralizing antibodies to HIV-1 induced by immunization. J Exp Med 210:209–223. https://doi.org/10.1084/jem.20121827
McFeeters RL, Xiong C, O’Keefe BR, Bokesch HR, McMahon JB, Ratner DM, Castelli R, Seeberger PH, Byrd RA (2007) The novel fold of scytovirin reveals a new twist for antiviral entry inhibitors. J Mol Biol 369:451–461. https://doi.org/10.1016/j.jmb.2007.03.030
Mir-Artigues P, Twyman RM, Alvarez D, Cerda Bennasser P, Balcells M, Christou P, Capell T (2019) A simplified techno-economic model for the molecular pharming of antibodies. Biotechnol Bioeng 116:2526–2539. https://doi.org/10.1002/bit.27093
Mitchell CA, Ramessar K, O’Keefe BR (2017) Antiviral lectins: selective inhibitors of viral entry. Antiviral Res 142:37–54. https://doi.org/10.1016/j.antiviral.2017.03.007
Mori T, Barrientos LG, Han Z, Gronenborn AM, Turpin JA, Boyd MR (2002) Functional homologs of cyanovirin-N amenable to mass production in prokaryotic and eukaryotic hosts. Protein Expr Purif 26:42–49. https://doi.org/10.1016/s1046-5928(02)00513-2
Mori T, O’Keefe BR, Sowder RC, Bringans S, Gardella R, Berg S, Cochran P, Turpin JA, Buckheit RW, McMahon JB (2005) Isolation and characterization of griffithsin, a novel HIV-inactivating protein, from the red alga Griffithsia sp. J Biol Chem 280:9345–9353. https://doi.org/10.1074/jbc.M411122200
Moulaei T, Botos I, Ziółkowska NE, Bokesch HR, Krumpe LR, McKee TC, O’Keefe BR, Dauter Z, Wlodawer A (2007) Atomic-resolution crystal structure of the antiviral lectin scytovirin. Protein Sci 16:2756–2760. https://doi.org/10.1110/ps.073157507
Moulaei T, Shenoy SR, Giomarelli B, Thomas C, McMahon JB, Dauter Z, O’Keefe BR, Wlodawer A (2010) Monomerization of viral entry inhibitor griffithsin elucidates the relationship between multivalent binding to carbohydrates and anti-HIV activity. Structure 18:1104–1115. https://doi.org/10.1016/j.str.2010.05.016
Naqvi S, Zhu C, Farre G, Ramessar K, Bassie L, Breitenbach J, Conesa DP, Ros G, Sandmann G, Capell T (2009) Transgenic multivitamin corn through biofortification of endosperm with three vitamins representing three distinct metabolic pathways. Proc Natl Acad Sci 106:7762–7767. https://doi.org/10.1073/pnas.0901412106
O’Keefe BR, Shenoy SR, Xie D, Zhang W, Muschik JM, Currens MJ, Chaiken I, Boyd MR (2000) Analysis of the interaction between the HIV-inactivating protein cyanovirin-N and soluble forms of the envelope glycoproteins gp120 and gp41. Mol Pharmacol 58:982–992. https://doi.org/10.1124/mol.58.5.982
O’Keefe BR, Vojdani F, Buffa V, Shattock RJ, Montefiori DC, Bakke J, Mirsalis J, d’Andrea A-L, Hume SD, Bratcher B (2009) Scaleable manufacture of HIV-1 entry inhibitor griffithsin and validation of its safety and efficacy as a topical microbicide component. Proc Natl Acad Sci 106:6099–6104. https://doi.org/10.1073/pnas.0901506106
O’Keefe BR, Giomarelli B, Barnard DL, Shenoy SR, Chan PKS, McMahon JB, Palmer KE, Barnett BW, Meyerholz DK, Wohlford-Lenane CL (2010) Broad-spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J Virol 84:2511–2521. https://doi.org/10.1128/JVI.02322-09
Ramessar K, Sabalza M, Capell T, Christou P (2008) Maize plants: an ideal production platform for effective and safe molecular pharming. Plant Sci 174:409–419. https://doi.org/10.1016/j.plantsci.2008.02.002
Ramessar K, Sabalza M, Miralpeix B, Capell T, Christou P (2010) Can microbicides turn the tide against HIV? Curr Pharm Des 16:468–485. https://doi.org/10.2174/138161210790232202
Ratner DM, Seeberger PH (2007) Carbohydrate microarrays as tools in HIV glycobiology. Curr Pharm Des 13:173–183. https://doi.org/10.2174/138161207779313650
Ratner D, Plante O, Seeberger P (2002) A linear synthesis of branched high-mannose oligosaccharides from the HIV-1 viral surface envelope glycoprotein gp120. Eur J Org Chem 2002:826–833. https://doi.org/10.1002/1099-0690(200203)2002:5%3c826::AID-EJOC826%3e3.0.CO;2-2
Sabalza M, Vamvaka E, Christou P, Capell T (2013) Seeds as a production system for molecular pharming applications: status and prospects. Curr Pharm Des 19:5543–5552. https://doi.org/10.2174/1381612811319310009
Sabalza M, Christou P, Capell T (2014) Recombinant plant-derived pharmaceutical proteins: current technical and economic bottlenecks. Biotechnol Lett 36:2367–2379. https://doi.org/10.1007/s10529-014-1621-3
Sacchettini JC, Baum LG, Brewer CF (2001) Multivalent protein–carbohydrate interactions. A new paradigm for supermolecular assembly and signal transduction. Biochemistry 40:3009–3015. https://doi.org/10.1021/bi002544j
Sánchez-Palomino S, Massanella M, Carrillo J, García A, García F, González N, Merino A, Alcamí J, Bofill M, Yuste E (2011) A cell-to-cell HIV transfer assay identifies humoral responses with broad neutralization activity. Vaccine 29:5250–5259. https://doi.org/10.1016/j.vaccine.2011.05.016
Shenoy SR, Barrientos LG, Ratner DM, O’Keefe BR, Seeberger PH, Gronenborn AM, Boyd MR (2002) Multisite and multivalent binding between cyanovirin-N and branched oligomannosides: calorimetric and NMR characterization. Chem Biol 10:1109–1118. https://doi.org/10.1016/s1074-5521(02)00237-5
Singh RS, Walia AK, Khattar JS, Singh DP, Kennedy JF (2017) Cyanobacterial lectins characteristics and their role as antiviral agents. Int J Biol Macromol 102:475–496. https://doi.org/10.1016/j.ijbiomac.2017.04.041
Siqueira AS, Lima ARJ, de Souza RC, Santos AS, da Vianez JLSG, Gonçalves EC, (2017) Anti-dengue virus activity of scytovirin and evaluation of point mutation effects by molecular dynamics and binding free energy calculations. Biochem Biophys Res Commun 490:1033–1038. https://doi.org/10.1016/j.bbrc.2017.06.160
Steckbeck JD, Kuhlmann AS, Montelaro RC (2013) C-terminal tail of human immunodeficiency virus gp41: functionally rich and structurally enigmatic. J Gen Virol 94:1–19. https://doi.org/10.1099/vir.0.046508-0
Stoger E, Ma JK-C, Fischer R, Christou P (2005) Sowing the seeds of success: pharmaceutical proteins from plants. Curr Opin Biotechnol 16:167–173. https://doi.org/10.1016/j.copbio.2005.01.005
Takebe Y, Saucedo CJ, Lund G, Uenishi R, Hase S, Tsuchiura T, Kneteman N, Ramessar K, Tyrrell DL, Shirakura M, Wakita T, McMahon JB, O’Keefe BR (2013) Antiviral lectins from red and blue-green algae show potent in vitro and in vivo activity against hepatitis C virus. PLoS ONE 8(5):e64449. https://doi.org/10.1371/journal.pone.0064449
Vamvaka E, Twyman RM, Murad AM, Melnik S, Teh AY, Arcalis E, Altmann F, Stoger E, Rech E, Ma JKC (2016a) Rice endosperm produces an underglycosylated and potent form of the HIV-neutralizing monoclonal antibody 2G12. Plant Biotechnol J 14:97–108. https://doi.org/10.1111/pbi.12360
Vamvaka E, Arcalis E, Ramessar K, Evans A, O’Keefe BR, Shattock RJ, Medina V, Stöger E, Christou P, Capell T (2016b) Rice endosperm is cost-effective for the production of recombinant griffithsin with potent activity against HIV. Plant Biotechnol J 14:1427–1437. https://doi.org/10.1111/pbi.12507
Vamvaka E, Evans A, Ramessar K, Krumpe LRH, Shattock RJ, O’Keefe BR, Christou P, Capell T (2016c) Cyanovirin-N produced in rice endosperm offers effective pre-exposure prophylaxis against HIV-1 BaL infection in vitro. Plant Cell Rep 35:1309–1319. https://doi.org/10.1007/s00299-016-1963-5
Vamvaka E, Farré G, Molinos-Albert LM, Evans A, Canela-Xandri A, Twyman RM, Ordóñez RACJ, Shattock RJ, O’Keefe BR, Clotet B, Blanco J, Khush GS, Christou P, Capell T (2018) Unexpected synergistic HIV neutralization by a triple microbicide produced in rice endosperm. Proc Natl Acad Sci 115:7854–7862. https://doi.org/10.1073/pnas.1806022115
World Health Organization (2020) HIV/AIDS. Data and statistics. Retrieved from https://www.who.int/hiv/data/en (Accesseed on 18 Nov 2020)
Xiong C, O’Keefe BR, Byrd RA, McMahon JB (2006) Potent anti-HIV activity of scytovirin domain 1 peptide. Peptides 27:1668–1675. https://doi.org/10.1016/j.peptides.2006.03.018
Acknowledgements
We acknowledge funding from Agencia de Gestio d’Ajuts Universitaris i de Recerca (AGAUR), Departament d’Empresa i Coneixement de la Generalitat de Catalunya (PANDEMIES 2020), MINECO, Spain (AGL2017-85377-R to T. Capell), Generalitat de Catalunya Grant 2017 SGR 828 to the Agricultural Biotechnology and Bioeconomy Unit (ABBU), CERCA Programme/Generalitat de Catalunya 2017 SGR 252 to IrsiCaixa, EU Pharma-Factory grant agreement 774078 to P. Christou and J. Blanco. A. Blanco-Perera was supported by a UdL fellowship. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261201800001I.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. This work was supported by Agencia de Gestio d’Ajuts Universitaris i de Recerca (AGAUR), Departament d’Empresa i Coneixement de la Generalitat de Catalunya (PANDEMIES 2020), MINECO, Spain (AGL2017-85377-R to T. Capell), Generalitat de Catalunya Grant 2017 SGR 828 to the Agricultural Biotechnology and Bioeconomy Unit (ABBU), CERCA Programme/Generalitat de Catalunya 2017 SGR 252 to IrsiCaixa, EU Pharma-Factory grant agreement 774078 to P. Christou and J. Blanco. A. Blanco-Perera was supported by a UdL fellowship. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261201800001I.
Author information
Authors and Affiliations
Contributions
VA-N, TC, BOK and PC: conceived and designed the research. VA-N, AB-P, SRS, YS and SM: conducted the experiments. JB, NI-U and BOK: commented critically on the manuscript and contributed with materials and expertise. VA-N, JM-B, DP-Z, JB, NI-U, BOK and PC: analyzed data and all co-authors contributed to the writing of the manuscript. All co-authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Stefan Schillberg.
The manuscript is dedicated to the memory of Ms. Yi Sun
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Armario-Najera, V., Blanco-Perera, A., Shenoy, S.R. et al. Physicochemical characterization of the recombinant lectin scytovirin and microbicidal activity of the SD1 domain produced in rice against HIV-1. Plant Cell Rep 41, 1013–1023 (2022). https://doi.org/10.1007/s00299-022-02834-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-022-02834-5