Abstract
Key message
Small RNAs have emerged as key players of gene expression regulation. Several lines of evidences highlight their role in modulating high temperature stress responsiveness in plants.
Abstract
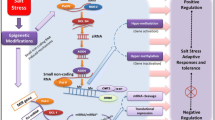
Throughout their life cycle, plants have to regulate their gene expression at various developmental phases, physiological changes, and in response to biotic or environmental stress. High temperature is one the most common abiotic stress for crop plants, that results in impaired morphology, physiology, and yield. However, plants have certain mechanisms that enable them to withstand such conditions by modulating the expression of stress-related genes. Small RNA (sRNA)-regulated gene expression is one such mechanism which is ubiquitous in all eukaryotes. The sRNAs mainly include micro RNAs (miRNAs) and small interfering RNAs (siRNAs). They are primarily associated with the gene silencing either through translation inhibition, mRNA degradation, or DNA methylation. During high temperature stress the increased or decreased level of miRNAs altered the protein accumulation of target transcripts and, therefore, regulate stress responses. Several reports are available in plants which are genetically engineered through expressing artificial miRNAs resulted in thermotolerance. sRNAs have also been reported to bring the epigenetic changes on chromatin region through RNA-dependent DNA methylation (RdDM). The present article draws a brief illustration of sRNA origin, their functional mechanisms, role in high temperature stress, and possible application for developing stress tolerant crop plants.


Similar content being viewed by others
References
Axtell MJ (2013) Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64:137–159
Axtell MJ, Jan C, Rajagopalan R, Bartel DP (2006) A two-hit trigger for siRNA biogenesis in plants. Cell 127:565–577
Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102:11928–11933
Bilichak A, Ilnytskyy Y, Woycicki R et al (2015) The elucidation of stress memory inheritance in Brassica rapa plants. Front Plant Sci 6:5
Blevins T, Podicheti R, Mishra V et al (2015) Identification of pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in arabidopsis. Elife 4:e09591
Bologna NG, Voinnet O (2014) The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol 65:473–503
Carbonell A, Daròs JA (2019) Design, synthesis, and functional analysis of highly specific artificial small RNAs with antiviral activity in plants. Methods Mol Biol 2028:231–246
Carbonell A, Takeda A, Fahlgren N et al (2014) New generation of artificial microRNA and synthetic trans-acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant Physiol 165:15–29
Carbonell A, Lisón P, Daròs JA (2019) Multi-targeting of viral RNAs with synthetic trans-acting small interfering RNAs enhances plant antiviral resistance. Plant J 100:720–737
Cisneros AE, Carbonell A (2020) Artificial small RNA-based silencing tools for antiviral resistance in plants. Plants 9:669
de Felippes FF, Marchais A, Sarazin A et al (2017) A single miR390 targeting event is sufficient for triggering TAS3-tasiRNA biogenesis in Arabidopsis. Nucleic Acids Res 45:5539–5554
de Jongh RPH, van Dijk ADJ, Julsing MK et al (2020) Designing eukaryotic gene expression regulation using machine learning. Trends Biotechnol 38:191–201
Ding Y, Ma Y, Liu N et al (2017) microRNAs involved in auxin signalling modulate male sterility under high-temperature stress in cotton (Gossypium hirsutum). Plant J 91:977–994
Fujimori S, Hasegawa T, Krey V et al (2019) A multi-model assessment of food security implications of climate change mitigation. Nat Sustain 2:386–396
Giacomelli JI, Weigel D, Chan RL, Manavella PA (2012) Role of recently evolved miRNA regulation of sunflower HaWRKY6 in response to temperature damage. New Phytol 195:766–773
Gottlieb Y, Zchori-Fein E, Mozes-Daube N et al (2010) The transmission efficiency of tomato yellow leaf curl virus by the whitefly Bemisia tabaci is correlated with the presence of a specific symbiotic bacterium species. J Virol 84:9310–9317
Guan Q, Lu X, Zeng H et al (2013) Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J 8:e24952
Hajheidari M, Farrona S, Huettel B et al (2012) CDKF;1 and CDKD protein kinases regulate phosphorylation of serine residues in the C-Terminal domain of Arabidopsis RNA polymerase II. Plant Cell 24:1626–1642
Halford NG, Curtis TY, Chen Z, Huang J (2015) Effects of abiotic stress and crop management on cereal grain composition: implications for food quality and safety. J Exp Bot 66:1145–1156
Iborra FJ, Jackson DA, Cook PR (2001) Coupled transcription and translation within nuclei of mammalian cells. Science 293:1139–1142
Kim YJ, Zheng B, Yu Y et al (2011) The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J 30:814–822
Kushawaha AK, Khan A, Sopory SK, Sanan-Mishra N (2021) Priming by high temperature stress induces microRNA regulated heat shock modules indicating their involvement in thermopriming response in rice. Life (basel) 11:291
Lai YS, Zhang X, Zhang W, Shen D, Wang H, Xia Y, Qiu Y, Song J, Wang C, Li X (2017) The association of changes in DNA methylation with temperature-dependent sex determination in cucumber. J Exp Bot 68:2899–2912
Li S, Liu J, Liu Z et al (2014) HEAT-INDUCED TAS1 TARGET1 mediates thermotolerance via heat stress transcription factor A1a-directed pathways in arabidopsis. Plant Cell 26:764–1780
Li H, Wang Y, Wang Z et al (2016) Microarray and genetic analysis reveals that csa-miR159b plays a critical role in abscisic acid-mediated heat tolerance in grafted cucumber plants. Plant Cell Environ 39:1790–1804
Li Y, Li X, Yang J, He Y (2020) Natural antisense transcripts of MIR398 genes suppress microR398 processing and attenuate plant thermotolerance. Nat Commun 11:5351
Lin JS, Kuo CC, Yang IC et al (2018) MicroRNA160 modulates plant development and heat shock protein gene expression to mediate heat tolerance in Arabidopsis. Front Plant Sci 9:68
Liu J, He Z (2020) Small DNA methylation, big player in plant abiotic stress responses and memory. Front Plant Sci 11:e595603
Liu J, Feng L, Li J, He Z (2015) Genetic and epigenetic control of plant heat responses. Front Plant Sci 6:267
Liu Y, Teng C, Xia R, Meyers BC (2020) PhasiRNAs in plants: their Biogenesis, genic sources, and roles in stress responses, development, and reproduction. Plant Cell 32:3059–3080
Malabarba J, Windels D, Xu W, Verdier J (2021) Regulation of DNA (de)methylation positively impacts seed germination during seed development under heat stress. Gene 12:457
Mao Y, Xu J, Wang Q et al (2021) A natural antisense transcript acts as a negative regulator for the maize drought stress response gene ZmNAC48. J Exp Bot 72:2790–2806
Matthews C, Arshad M, Hannoufa A (2019) Alfalfa response to heat stress is modulated by microRNA156. Physiol Plant 165:830–842
Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15:394–408
Naydenov M, Baev V, Apostolova E et al (2015) High-temperature effect on genes engaged in DNA methylation and affected by DNA methylation in Arabidopsis. Plant Physiol Biochem 87:102–108
Papareddy RK, Páldi K, Paulraj S, Kao P, Lutzmayer S, Nodine MD (2020) Chromatin regulates expression of small RNAs to help maintain transposon methylome homeostasis in Arabidopsis. Genome Biol 21:251
Patel P, Yadav K, Srivastava AK et al (2019) Overexpression of native Musa-miR397 enhances plant biomass without compromising abiotic stress tolerance in banana. Sci Rep 9:e16434
Popova OV, Dinh HQ, Aufsatz W, Jonak C (2013) The RdDM pathway is required for basal heat tolerance in arabidopsis. Mol Plant 6:396–410
Prasad A, Sharma N, Muthamilarasan M et al (2019) Recent advances in small RNA mediated plant-virus interactions. Crit Rev Biotechnol 39:587–601
Prasad A, Sharma N, Prasad M (2020) Noncoding but CODING: Pri-miRNA into the action. Trends Plant Sci 26:204–206
Ravichandran S, Ragupathy R, Edwards T, Domaratzki M, Cloutier S (2019) MicroRNA-guided regulation of heat stress response in wheat. BMC Genomics 20:488
Sharma N, Prasad M (2020) Silencing AC1 of Tomato leaf curl virus using artificial microRNA confers resistance to leaf curl disease in transgenic tomato. Plant Cell Rep 39:1565–1579
Shi X, Jiang F, Wen J, Wu Z (2019) Overexpression of Solanum habrochaites microRNA319d (sha-miR319d) confers chilling and heat stress tolerance in tomato (S. lycopersicum). BMC Plant Biol 19:214
Singh RK, Prasad M (2021) Delineating the epigenetic regulation of heat and drought response in plants. Crit Rev Biotechnol. https://doi.org/10.1080/07388551.2021.1946004
Singh A, Gautam V, Singh S et al (2018) Plant small RNAs: advancement in the understanding of biogenesis and role in plant development. Planta 248:545–558
Singh RK, Prasad A, Muthamilarasan M, Parida SK, Prasad M (2020) Breeding and biotechnological interventions for trait improvement: status and prospects. Planta 252:54
Stief A, Altmann S, Hoffmann K et al (2014) Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26:1792–1807
Szaker HM, Darkó É, Medzihradszky A et al (2019) miR824/AGAMOUS-LIKE16 module integrates recurring environmental heat stress changes to fine-tune poststress development. Front Plant Sci 10:1454
Varkonyi-Gasic E, Wu R, Wood M et al (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3:1–12
Vazquez F, Gasciolli V, Crété P, Vaucheret H (2004) The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 14:346–351
Wang F, Axtell MJ (2017) AGO4 is specifically required for heterochromatic siRNA accumulation at Pol V-dependent loci in Arabidopsis thaliana. Plant J 90:37–47
Wang XJ, Gaasterland T, Chua NH (2005) Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol 6:R30
Wang Y, Sun F, Cao H et al (2012) TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS ONE 7:e48445
Yan J, Gu Y, Jia X et al (2012) Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24:415–427
Yang L, Liu Z, Lu F et al (2006) SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J 47:841–850
Yu B, Yang Z, Li J et al (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307:932–935
Yu R, Jih G, Iglesias N, Moazed D (2014) Determinants of heterochromatic siRNA biogenesis and function. Mol Cell 53:262–276
Yuan C, Wang J, Harrison AP et al (2014) Genome-wide view of natural antisense transcripts in Arabidopsis thaliana. DNA Res 22:233–243
Zhang X, Lii Y, Wu Z et al (2013) Mechanisms of small RNA generation from Cis-NATs in response to environmental and developmental cues. Mol Plant 6:704–715
Zhao J, Yuan S, Zhou M et al (2019) Transgenic creeping bentgrass overexpressing Osa-miR393a exhibits altered plant development and improved multiple stress tolerance. Plant Biotechnol J 17:233–251
Zhao Y, Xie J, Wang S et al (2021) Synonymous mutation of miR396a target sites in Growth Regulating Factor 15 (GRF15) enhances photosynthetic efficiency and heat tolerance in poplar. J Exp Bot. https://doi.org/10.1093/jxb/erab120
Acknowledgements
Authors’ work in this area is supported by J.C. Bose National Fellowship Grant of Department of Science and Technology [JCB/2018/000001] and core grant of DBT-NIPGR. RKS acknowledges the DBT Multi-institutional project entitled—“Germplasm Characterization and Trait Discovery in Wheat using Genomics Approaches and its Integration for Improving Climate Resilience, Productivity and Nutritional quality" under mission programme of "Characterisation of Genetic Resources”, [BT/Ag/Network/Wheat/2019-20] for the research grant. AP and JM acknowledge the research fellowship from Council of Scientific and Industrial Research, Govt. of India. The authors are thankful to DBT-eLibrary Consortium (DeLCON) for providing access to e-resources.
Author information
Authors and Affiliations
Contributions
MP conceived the idea and outlined the review. RKS, AP, and JM prepared the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest declared.
Additional information
Communicated by Vijay Pratap Singh.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Singh, R.K., Prasad, A., Maurya, J. et al. Regulation of small RNA-mediated high temperature stress responses in crop plants. Plant Cell Rep 41, 765–773 (2022). https://doi.org/10.1007/s00299-021-02745-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-021-02745-x