Abstract
Key message
Cold-tolerance in rice may be related to increased cellulose deposition in the cell wall, membrane fatty acids unsaturation and differential expression of several newly identified genes.
Abstract
Low temperature exposure during early vegetative stages limits rice plant’s growth and development. Most genes previously related to cold tolerance in rice are from the japonica subspecies. To help clarify the mechanisms that regulate cold tolerance in young indica rice plants, comparative transcriptome analysis of 6 h cold-treated (10 °C) leaves from two genotypes, cold-tolerant (CT) and cold-sensitive (CS), was performed. Differentially expressed genes were identified: 831 and 357 sequences more expressed in the tolerant and in the sensitive genotype, respectively. The genes with higher expression in the CT genotype were used in systems biology analyses to identify protein–protein interaction (PPI) networks and nodes (proteins) that are hubs and bottlenecks in the PPI. From the genes more expressed in the tolerant plants, 60% were reported as affected by cold in previous transcriptome experiments and 27% are located within QTLs related to cold tolerance during the vegetative stage. Novel cold-responsive genes were identified. Quantitative RT-PCR confirmed the high-quality of RNAseq libraries. Several genes related to cell wall assembly or reinforcement are cold-induced or constitutively highly expressed in the tolerant genotype. Cold-tolerant plants have increased cellulose deposition under cold. Genes related to lipid metabolism are more expressed in the tolerant genotype, which has higher membrane fatty acids unsaturation, with increasing levels of linoleic acid under cold. The CT genotype seems to have higher photosynthetic efficiency and antioxidant capacity, as well as more effective ethylene, Ca2+ and hormone signaling than the CS. These genes could be useful in future biotechnological approaches aiming to increase cold tolerance in rice.









Similar content being viewed by others
References
Aalto MK, Helenius E, Kariola T, Pennanen V, Heino P, Hõrak H, Puzõrjova I, Kollist H, Palva ET (2012) ERD15—an attenuator of plant ABA responses and stomatal aperture. Plant Sci 182:19–28
Abuqamar S, Ajeb S, Sham A, Enan MR, Iratni R (2013) A mutation in the expansin-like A2 gene enhances resistance to necrotrophic fungi and hypersensitivity to abiotic stress in Arabidopsis thaliana. Mol Plant Pathol 14:813–827
Adamski JM, Cargnelutti D, Sperotto RA, Terra TF, Rosa LMG, Cruz RP, Fett JP (2016) Identification and physiological characterization of two sister lines of indica rice (Oryza sativa L.) with contrasting levels of cold tolerance. Can J Plant Sci 96:197–214
Aguan K, Sugawara K, Susuki N, Kusano T (1991) Isolation of genes for low-temperature-induced proteins in rice by a simple subtractive method. Plant Cell Physiol 32:1285–1289
Amaral MN, Arge LWP, Benitez LC, Danielowski R, Silveira SFS, Farias DR, Oliveira AC, Maia LC, Braga EJB (2016) Comparative transcriptomics of rice plants under cold, iron, and salt stresses. Funct Integr Genomics 16:567–579
Andaya VC, McKill DJ (2003) Mapping of QTL associated with cold tolerance during the vegetative stage in rice. J Exp Bot 54:2579–2585
Andaya VC, Tai TH (2006) Fine mapping of the qCTS12 locus, a major QTL for seedling cold tolerance in rice. Theor Appl Genet 113:467–475
Assenov Y, Ramírez F, Schelhorn S-E, Lengauer T, Albrecht M (2008) Computing topological parameters of biological networks. Bioinformatics 24(2):282–284
Bader GD, Hogue CW (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform 4:2
Barrett T, Wilhite SE, Ledoux P et al (2013) NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41(Database issue):D991–D995. https://doi.org/10.1093/nar/gks1193
Bevilacqua CB, Basu S, Pereira A, Tseng TM, Zimmer PD, Burgos NR (2015) Analysis of stress-responsive gene expression in cultivated and weedy rice differing in cold stress tolerance. PLoS One 10:e0132100
Bligh EC, Dyer WJ (1959) A rapid method of total lipid. Extraction and purification. Can J Biochem Physiol 37:911–917
Bonnecarrère V, Borsani O, Díaz P, Capdevielle F, Blanco P, Monza J (2011) Response to photoxidative stress induced by cold in japonica rice is genotype dependent. Plant Sci 180:726–732
Breton G, Danyluk J, Charron J, Sarhan F (2003) Expression profiling and bioinformatic analyses of a novel stress-regulated multispanning transmembrane protein family from cereals and Arabidopsis. Plant Physiol 32:64–74
Butelli E, Licciardello C, Zhang Y, Liu J, Mackay S, Bailey P, Reforgiato-Recupero G, Martin C (2012) Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 3:1242–1255
Chandran AKN, Jeong HY, Jung K-H, Lee C (2016) Development of functional modules based on co-expression patterns for cell-wall biosynthesis related genes in rice. J Plant Biol 59:1–15
Chawade A, Lindlöf A, Olsson B, Olsson O (2013) Global expression profiling of low temperature induced genes in the chilling tolerant japonica rice Jumli Marshi. PLoS One 8:e81729
Chen NA, Xu Y, Wang X, Du C, Du J, Yuan M, Xu Z, Chong K (2011) OsRAN2, essential for mitosis, enhances cold tolerance in rice by promoting export of intranuclear tubulin and maintaining cell division under cold stress. Plant Cell Environ 34:52–64
Cheng C, Yun KY, Ressom HW, Mohanty B, Bajic VB, Jia Y, Yun SJ, de los Reyes BG (2007) An early response regulatory cluster induced by low temperature and hydrogen peroxide in seedlings of chilling-tolerant japonica rice. BMC Genom 8:175
Cheng YW, Feng AQ, Zhang ZB, Zhang YZ, Han JM (2014) The phylogenetic analysis and identification of a novel remorin member from rice (Oryza sativa L) by proteomics under salt stress. Adv Mat Res 1073–1076:229–232
Chinnusamy V, Zhu JK, Sunkar R (2010) Gene regulation during cold stress acclimation in plants. Methods Mol Biol 639:39–55
Colebrook EH, Thomas SG, Phillips AL, Hedden P (2014) The role of gibberellin signaling in plant responses to abiotic stress. J Exp Biol 217:67–75
Conesa A, Götz A (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genom 2008:619832
Copetti D, Zhang J, El Baidouri M, Gao D, Wang J, Barghini E, Cossu RM, Angelova A, Maldonado LCE, Roffler S, Ohyanagi H, Wicker T, Fan C, Zuccolo A, Chen M, de Oliveira AC, Han B, Henry R, Hsing Y-I, Kurata N, Wang W, Jackson SA, Panaud O, Wing RA (2015) RiTE database: a resource database for genus-wide rice genomics and evolutionary biology. BMC Genom 16:538
Cruz RP, Milach SCK (2004) Cold tolerance at the germination stage of rice: methods of evaluation and characterization of genotypes. Sci Agric 61:1–8
Cruz RP, Golombieski JI, Bazana MT, Cabreira C, Silveira TF, da Silva LP (2010) Alterations in fatty acid composition due to cold exposure at the vegetative stage in rice. Braz J Plant Physiol 22:199–207
Cruz RP, Sperotto RA, Cargnelutti D, Adamski JM, Terra TF, Fett JP (2013) Avoiding damage and achieving cold tolerance in rice plants. Food Energy Secur 2:96–119
Dametto A, Sperotto RA, Adamski JM, Blasi EA, Cargnelutti D, de Oliveira LF, Ricachenevsky FK, Fregonezi JN, Mariath JE, da Cruz RP, Margis R, Fett JP (2015) Cold tolerance in rice germinating seeds revealed by deep RNAseq analysis of contrasting indica genotypes. Plant Sci 238:1–12
Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought, high-salt- and cold-responsive gene expression. Plant J 33:751–763
El Baidouri M, Panaud O (2013) Comparative genomic paleontology across plant kingdom reveals the dynamics of TE-driven genome evolution. Genome Biol Evol 5:954–965
Endler A, Kesten C, Schneider R, Zhang Y, Ivakov A, Froehlich A, Funke N, Persson S (2015) A mechanism for sustained cellulose synthesis during salt stress. Cell 162:1353–1364
Endler A, Schneider R, Kesten C, Lampugnani ER, Persson S (2016) The cellulose synthase companion proteins act non-redundantly with CELLULOSE SYNTHASE INTERACTING1/POM2 and CELLULOSE SYNTHASE 6. Plant Signal Behav 11(4):e1135281
Fang H, Meng Q, Xu J, Tang H, Tang S, Zhang H, Huang J (2015) Knock-down of stress inducible OsSRFP1 encoding an E3 ubiquitin ligase with transcriptional activation activity confers abiotic stress tolerance through enhancing antioxidant protection in rice. Plant Mol Biol 87:441–458
FinattoT, De Oliveira AC, Chaparro C, Da Maia LC, Farias DR, Woyann LG, etal (2015). Abiotic stress and genome dynamics: specific genes and transposable elements response to iron excess in rice. Rice 8:1. https://doi.org/10.1186/s12284-015-0045-6
Frenette Charron JB, Breton G, Badawi M, Sarhan F (2002) Molecular and structural analyses of a novel temperature stress-induced lipocalin from wheat and Arabidopsis. FEBS Lett 517:129–132
Gothandam KM, Nalini E, Karthikeyan S, Shin JS (2010) OsPRP3, a flower specific proline-rich protein of rice, determines extracellular matrix structure of floral organs and its overexpression confers cold-tolerance. Plant Mol Biol 72:125–135
Guo-Li W, Zhen-Fei G (2005) Effects of chilling stress on photosynthetic rate and chlorophyll fluorescence parameter in seedlings of two rice cultivars differing in cold tolerance. Rice Sci 12:187–191
Hartman L, Lago BCA (1973) Rapid preparation of fatty, methyl esters from lipids. Lab Pract 22:457–477
Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI (2001) U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem 276(35):33111–33120
He ZH, Dong HT, Dong JX, Li DB, Ronald PC (2000) The rice Rim2 transcript accumulates in response to Magnaporthe grisea and its predicted protein product shares similarity with TNP2- like proteins encoded by CACTA transposons. Mol Gen Genet 264:2–10
Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992
Huang J, Sun S, Xu D, Lan H, Sun H, Wang Z, Bao Y, Wang J, Tang H, Zhang H (2012) A TFIIIA-type zinc finger protein confers multiple abiotic stress tolerances in transgenic rice (Oryza sativa L.). Plant Mol Biol 80:337–350
Huang F, Lian L, He W, Zhu Y, Cai Q, Xie H, Zhang J (2014a) Genome-wide profiling of changes in gene expression in response to infection of the japonica rice variety Yunyin by Magnaporthe oryzae. Mol Breed 34:1965–1974
Huang L, Zhang F, Zhang F, Wang W, Zhou Y, Fu B, Li Z (2014b) Comparative transcriptome sequencing of tolerant rice introgression line and its parents in response to drought stress. BMC Genom 15:1026
Huang L, Hong Y, Zhang H, Li D, Song F (2016) Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol 16:203
Iba K (2002) Acclimative response to temperature stress in higher plants: approaches of gene engineering for temperature tolerance. Annu Rev Plant Biol 53:225–245
Inagaki Y-S, Ethrerington G, Geisler K, Field B, Dokarry M, Ikeda K, Mutsukado Y, Dicks J, Osbourn A (2011) Investigation of the potential for triterpene synthesis in rice through genome mining and metabolic engineering. New Phytol 191:432–448
IRGSP-International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Ishiguro S, Ogasawara K, Fujino K, Sato Y, Kishima Y (2014) Low temperature-responsive changes in the anther transcriptome’s repeat sequences are indicative of stress sensitivity and pollen sterility in rice strains. Plant Physiol 164:671–682. https://doi.org/10.1104/pp.113.230656
Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47:141–153
Jami SK, Clark GB, Ayele BT, Roux SJ, Kirti PB (2012) Identification and characterization of annexin gene family in rice. Plant Cell Rep 31:813–825
Jarsch IK, Ott T (2011) Perspectives on remorin proteins, membrane rafts, and their role during plant-microbe interactions. Mol Plant Microbe Interact 24:7–12
Jiang L, Xun M, Wang J, Wan J (2008) QTL analysis of cold tolerance at seedling stage in rice (Oryza sativa L.) using recombination inbred lines. J Cereal Sci 48:173–179
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Kargiotidou A, Deli D, Galanopolou D, Tsaftaris A, Farmaki T (2008) Low temperature and light regulate delta 12 fatty acid desaturases (FAD2) at a transcriptional level in cotton (Gossypium hirsutum). J Exp Bot 59:2043–2056
Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, Childs KL, Davidson RM, Lin H, Quesada-Ocampo L, Vaillancourt B, Sakai H, Lee SS, Kim J, Numa H, Itoh T, Buell CR, Matsumoto T (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4
Kim SJ, Lee SC, Hong SK, An K, An G, Kim SR (2009) Ectopic expression of a cold-responsive OsAsr1 cDNA gives enhanced cold tolerance in transgenic rice plants. Mol Cells 27:449–458
Kim SM, Suh JP, Lee CK, Lee JH, Kim YG, Jena KK (2014) QTL mapping and development of candidate gene-derived DNA markers associated with seedling cold tolerance in rice (Oryza sativa L.). Mol Genet Genom 289:333–343
Konopka-Postupolska D, Clark G, Hofmann A (2011) Structure, function and membrane interactions of plant annexins: an update. Plant Sci 181:230–241
Koseki M, Kitazawa N, Yonebayashi S, Maehara Y, Wang ZX, Minobe Y (2010) Identification and fine mapping of a major quantitative trait locus originating from wild rice, controlling cold tolerance at the seedling stage. Mol Genet Genom 284:45–54
Kumari S, Sabharwal VP, Kushwaha HR, Sopory SK, Singla-Pareek SL, Pareek A (2009) Transcriptome map for seedling stage specific salinity stress response indicates a specific set of genes as candidate for saline tolerance in Oryza sativa L. Funct Integr Genom 9:109–123
Kunze M, Pracharoenwattana I, Smith SM, Hartig A (2006) A central role for the peroxisomal membrane in glyoxylate cycle function. Biochim Biophys Acta 1763:1441–1452
Lee SC, Huh KW, An K, An G, Kim SR (2004a) Ectopic expression of a cold-inducible transcription factor, CBF1/DREB1b, in transgenic rice (Oryza sativa L.). Mol Cells 18:107–114
Lee S, Lee EJ, Yang EJ, Lee JE, Park AR, Song WH, Park OK (2004b) Proteomic identification of annexins, calcium-dependent membrane binding proteins that mediate osmotic stress and abscisic acid signal transduction in Arabidopsis. Plant Cell 16:1378–1391
Lee SC, Lee MY, Kim SJ, Jun SH, An G, Kim SR (2005) Characterization of an abiotic stress-inducible dehydrin gene, OsDhn1, in rice (Oryza sativa L.). Mol Cells 19:212–218
Lee J, Lee W, Kwon SW (2015) A quantitative shotgun proteomics analysis of germinated rice embryos and coleoptiles under low-temperature conditions. Proteome Sci 13:27
Li TG, Visperas RM, Vergara BS (1981) Correlation of cold tolerance at different growth stages in rice. Acta Bot Sin 23:203–207
Li Y, Qian Q, Zhou Y, Yan M, Sun L, Zhang M, Fu Z, Wang Y, Han B, Pang X, Chen M, Li J (2003) BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15:2020–2031
Lindlöf A, Chawade A, Sikora P, Olsson O (2015) Comparative transcriptomics of Sijung and Jumli Marshi rice during early chilling stress imply multiple protective mechanisms. PLoS One 10:e0125385
Liu CW, Hsu YK, Cheng YH, Yen HC, Wu YP, Wang CS, Lai CC (2012) Proteomic analysis of salt-responsive ubiquitin-related proteins in rice roots. Rapid Commun Mass Spectrom 26:1649–1660
Lou Q, Chen L, Sun Z, Xing Y, Li J, Xu X, Mei H, Luo L (2007) A major QTL associated with cold tolerance at seedling stage in rice (Oryza sativa L.). Euphytica 158:87–94
Lourenço T, Sapeta H, Figueiredo DD, Rodrigues M, Cordeiro A, Abreu IA, Saibo NJ, Oliveira MM (2013) Isolation and characterization of rice (Oryza sativa L.) E3-ubiquitin ligase OsHOS1 gene in the modulation of cold stress response. Plant Mol Biol 83:351–363
Ma Y, Dai X, Xu Y, Luo W, Zheng X, Zeng D, Pan Y, Lin X, Liu H, Zhang D, Xiao J, Guo X, Xu S, Niu Y, Jin J, Zhang H, Xu X, Li L, Wang W, Qian Q, Ge S, Chong K (2015) COLD1 confers chilling tolerance in rice. Cell 160:1209–1221
Mackill DJ, Lei X (1997) Genetic variation for traits related to temperate adaptation of rice cultivars. Crop Sci 37:1340–1346
Makarevitch I, Waters AJ, West PT, Stitzer M, Hirsch CN et al (2015) Transposable Elements Contribute to Activation of Maize Genes in Response to Abiotic Stress. PLoS Genet 11(1):e1004915. https://doi.org/10.1371/journal.pgen.1004915
Mertz LM, Henning FA, Soares RC, Baldiga RF, Peske FB, Moraes DM (2009) Physiological changes in rice seeds exposed to cold in the germination phase. Rev Bras Sem 31:254–262
Mishra M, Kanwar P, Singh A, Pandey A, Kapoor S, Pandey GK (2003) Plant Omics: genome-wide analysis of ABA Repressor1 (ABR1) related genes in rice during abiotic stress and development. OMICS 17(8): 439–450
Mittal D, Madhyastha DA, Grover A (2012) Genome-wide transcriptional profiles during temperature and oxidative stress reveal coordinated expression patterns and overlapping regulons in rice. PLoS One 7:e40899
Mizutani M (2012) Impacts of diversification of cytochrome P450 on plant metabolism. Biol Pharm Bull 35:824–832
Mori M, Onishi K, Tokizono Y, Shinada H, Yushimura T, Numao Y et al (2011) Detection of a novel quantitative trait loci for cold tolerance at the booting stage derived from a tropical japonica rice variety Silewah. Breed Sci 61:61–68
Murata N, Ishizaki-Nishizawa O, Higashi S, Hayashi H, Tasaka Y, Nishida I (1992) Genetically engineered alteration in the chilling sensitivity of plants. Nature 356:710–713
Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, Richardson AO, Okumoto Y, Tanisaka T, Wessler SR (2009) Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461:1130–1134
Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51:617–630
Negi P, Rai AN, Suprasanna P (2016) Moving through the stressed genome: emerging regulatory roles for transposons in plant stress response. Front Plant Sci 7:1448
Neumann P, Yan H, Jiang J (2007) The centromeric retrotransposons of rice are transcribed and differentially processed by RNA interference. Genetics 176:749–761. https://doi.org/10.1534/genetics.107.071902
Nie D-M, Ouyang Y-D, Wang X, Zhou W, Hu C-G, Yao J (2013) Genome-wide analysis of endosperm-specific genes in rice. Gene 530:236–247
O’Brien JA, Benková E (2013) Cytokinin cross-talking during biotic and abiotic stress responses. Front Plant Sci 4:451
Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran L-S P (2013) Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress. J Exp Bot 64:445–458
Pan Y, Zhang H, Zhang D, Li J, Xiong H, Yu J et al (2015) Genetic analysis of cold tolerance at the germination and booting stages in rice by association mapping. PLoS One 10:e0120590
Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133:1755–1767
Raffaele S, Mongrand S, Gamas P, Niebel A, Ott T (2007) Genome-wide annotation of remorins, a plant-specific protein family: evolutionary and functional perspectives. Plant Physiol 145:593–600
Rahman A (2013) Auxin: a regulator of cold stress response. Physiol Plant 147:28–35
Ranawake AL, Manangkil OE, Yoshida S, Ishii T, Mori N, Nakamura C (2014) Mapping QTLs for cold tolerance at germination and the early seedling stage in rice (Oryza sativa L.). Biotechnol Biotechnol Equip 28:989–998
Robinson MD, McCarthy DJ, Smyth GK (2010) EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Román A, Andreu V, Hernández ML, Lagunas B, Picorel R, Martínez-Rivas JM, Alfonso M (2012) Contribution of the different omega-3 fatty acid desaturase genes to the cold response in soybean. J Exp Bot 63:4973–4982
Routaboul JM, Fischer SF, Browse J (2000) Trienoic fatty acids are required to maintain chloroplast function at low temperatures. Plant Physiol 124:1697–1705
Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23:319–327
Saito KSY, Kuroki M, Sato Y (2010) Map-based cloning of the rice cold tolerance gene Ctb1. Plant Sci 179:97–102
Sakai H, Lee SS, Tanaka T, Numa H, Kim J, Kawahara Y, Wakimoto H, Yang CC, Iwamoto M, Abe T, Yamada Y, Muto A, Inokuchi H, Ikemura T, Matsumoto T, Sasaki T, Itoh T (2013) Rice annotation project database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol 54(2):e6
Sanghera GS, Wani SH, Hussain W, Singh NB (2011) Engineering cold stress tolerance in crop plants. Curr Genom 12:30–43
Sato Y, Masuta Y, Saito K, Murayama S, Ozawa K (2011) Enhanced chilling tolerance at the booting stage in rice by transgenic overexpression of the ascorbate peroxidase gene, OsAPXa. Plant Cell Rep 30:399–406
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108
Schuler MA, Werck-Reichhart D (2003) Functional Genomics of P450s. Annu Rev Plant Biol 54:629–667
Shakiba E, Edwards JD, Jodari F, Duke SE, Baldo AM, Korniliev P et al. (2017) Genetic architecture of cold tolerance in rice (Oryza sativa) determined through high resolution genome-wide analysis. PLoS One 12(3): e0172133. https://doi.org/10.1371/journal.pone.0172133
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Shi J, Cao Y, Fan X, Li M, Wang Y, Ming F (2012) A rice microsomal delta-12 fatty acid desaturase can enhance resistance to cold stress in yeast and Oryza sativa. Mol Breed 29:743–757
Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55:555–590
Song X, Cao X (2017) Transposon-mediated epigenetic regulation contributes to phenotypic diversity and environmental adaptaion in rice. Curr Opin Plant Biol 36:111–118
Songyikhangsuthor K, Guo Z, Wang N, Zhu X, Xie W, Mou T, Xiong L (2014) Natural variation in the sequence of SNAC1 and its expression level polymorphism in rice germplasms under drought stress. J Genet Genom 41:609–612
Strasser RJ, Tsimilli-Michael M, Srivastava A (2004) Chlorophyll a fluorescence: a signature of photosynthesis, analysis of the chlorophyll a fluorescence transient, vol 14. Springer, Dordrecht, pp 321–362 (Chapter 12)
Su CF, Wang YC, Hsieh TH, Lu CA, Tseng TH, Yu SM (2010) A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol 153:145–158
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:447–452
Takahashi N (1984) Differentiation of ecotypes in Oryza sativa. In: Takahashi N, Tsunoda S (eds) Biology of rice. Japan Science Society, Tokyo, pp 31–67
Tang W, Page M (2013) Transcription factor AtbZIP60 regulates expression of Ca2+-dependent protein kinase genes in transgenic cells. Mol Biol Rep 40:2723–2732
Tao Z, Kou Y, Liu H, Li X, Xiao J, Wang S (2011) OsWRKY45 alleles play different roles in abscisic acid signaling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J Exp Bot 62:4863–4874
Tian Y, Zhang H, Pan X, Chen X, Zhang Z, Lu X et al (2011) Overexpression of ethylene response factor TERF2 confers cold tolerance in rice seedlings. Transg Res 20:857–866
Tittel-Elmer M, Bucher E, Broger L, Mathieu O, Paszkowski J, Vaillant I et al (2010) Stress-induced activation of heterochromatic transcription. PLoS Genet 6:e1001175. https://doi.org/10.1371/journal.pgen.1001175
Tovuu A, Zulfugarov I, Wu G, Kang IS, Kim C, Moon BY, An G, Lee C-H (2016) Rice mutants deficient in ω-3 fatty acid desaturase (FAD8) fail to acclimate to cold temperatures. Plant Phys Biochem 109:525–535
Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M (2009) A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, maize. Plant Cell Environ 32:1211–1229
Valitova JN, Sulkarnayeva AG, Minibayeva FV (2016) Plant Sterols: diversity, biosynthesis, and physiological functions. Biochem (Moscow) 81:819–834
Vij S, Giri J, Dansana PK, Kapoor S, Tyagi AK (2008) The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: organization, phylogenetic relationship, and expression during development and stress. Mol Plant 1:732–750
Wang GD, Tian PF, Cheng Z-K, Wu ZK, Jiang G, Li JM, Li DB, He Q ZH (2003) Genomic characterization of Rim2 / Hipa elements reveals a CACTA-like transposon superfamily with unique features in the rice genome. Mol Gen Genom 270:234–242
Wang Q, Guan Y, Wu Y, Chen H, Chen F, Chu C (2008) Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol Biol 67:589–602
Wen JQ, Oono K, Imai R (2002) Two novel mitogen-activated protein signaling components, OsMEK1 and OsMAP1, are involved in a moderate low-temperature signaling pathway in rice. Plant Physiol 129:1880–1891
Werner T, Schmülling T (2009) Cytokinin action in plant development. Curr Opin Plant Biol 12:527–538
Winfield MO, Lu C, Wilson ID, Coghill JA, Edwards KJ (2010) Plant responses to cold: transcriptome analysis of wheat. Plant Biotech J 8:749–771
Wu Y-S, Yang C-Y (2016) Physiological responses and expression profile of NADPH oxidase in rice (Oryza Sativa) seedlings under different levels of submergence. Rice 9:2
Xie G, Kato H, Imai R (2012) Biochemical identification of the OsMKK6-OsMPK3 signaling pathway for chilling stress tolerance in rice. Biochem J 443:95–102
Xiong L, Yang Y (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15:745–759
Yamaguchi T, Nakayama K, Hayashi T, Yazaki J, Kishimoto N, Kikuchi S, Koike S (2004) cDNA microarray analysis of rice anther genes under chilling stress at the microsporogenesis stage revealed two genes with DNA transposon Castway in the 5′-flanking region. Biosci Biotechnol Biochem 68(6):1315–1323
Yan YS, Chen XY, Yang K, Sun ZX, Fu YP, Zhang YM, Fang RX (2011) Overexpression of an F-box protein gene reduces abiotic stress tolerance and promotes root growth in rice. Mol Plant 4:190–197
Yang YW, Chen HC, Jen WF, Liu LY, Chang MC (2015) Comparative transcriptome analysis of shoots and roots of TNG67 and TCN1 rice seedlings under cold stress and following subsequent recovery: insights into metabolic pathways, phytohormones, and transcription factors. PLoS One 10:e0131391
Ye H, Du H, Tang N, Li X, Xiong L (2009) Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol Biol 71:291–305
Yen SK, Chung MC, Chen PC, Yen HE (2001) Environmental and developmental regulation of the wound-induced cell wall protein WI12 in the halophyte ice plant. Plant Physiol 127:517–528
Zeller G, Henz SR, Widmer CK, Sachsenberg T, Rätsch G, Weigel D et al (2009) Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J 58:1068–1082. https://doi.org/10.1111/j.1365-313X.2009.03835.x
Zhang ZH, Qu XS, Wan S, Chen LH, Zhu YG (2005) Comparison of QTL controlling seedling vigour under different temperature conditions using recombinant inbred lines in rice (Oryza sativa). Ann Bot 95:423–429
Zhang T, Zhao X, Wang W, Pan Y, Huang L, Liu X et al (2012a) Comparative transcriptome profiling of chilling stress responsiveness in two contrasting rice genotypes. PLoS One 7:e43274
Zhang F, Huang L, Wang W, Zhao X, Zhu L, Fu B, Li Z (2012b) Genome-wide gene expression profiling of introgressed indica rice alleles associated with seedling cold tolerance improvement in a japonica rice background. BMC Genom 13:461
Zhang Q, Jiang N, Wang GL, Hong Y, Wang Z (2013) Advances in understanding cold sensing and the cold-responsive network in rice. Adv Crop Sci Technol 1:1
Zhang J, Luo W, Zhao Y, Xu Y, Song S, Chong K (2016) Comparative metabolomic analysis reveals a reactive oxygen species-dominated dynamic model underlying chilling environment adaptation and tolerance in rice. New Phytol 211:1295–1310. https://doi.org/10.1111/nph.14011
Zheng M, Wang Y, Liu K, Shu H, Zhou Z (2012) Protein expression changes during cotton fiber elongation in response to low temperature stress. J Plant Physiol 169:399–409
Zheng W, Ma L, Zhao J, Li Z, Sun F, Lu X (2013) Comparative transcriptome analysis of two rice varieties in response to rice stripe virus and small brown planthoppers during early interaction. PLoS One 8:e82126
Zhiguo E, Zhang Y, Li T, Wang L, Zhao H (2015) Characterization of the ubiquitin-conjugating enzyme gene family in rice and evaluation of expression profiles under abiotic stresses and hormone treatments. PLoS One 10(4):e0122621. https://doi.org/10.1371/journal.pone.0122621
Zhu JK (2016) Abiotic stress signaling and responses in plants. Cell 167:313–324
Zhu Y, Chen K, Mi X, Chen T, Ali J, Ye G et al (2015) Identification and fine mapping of a stably expressed QTL for cold tolerance at the booting stage using an interconnected breeding population in rice. PLoS One 10:e0145704
Acknowledgements
This work was supported by Universidade do Vale do Taquari - UNIVATES and the Brazilian funding agencies FAPERGS (Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico). The authors thank IRGA (Instituto Rio-Grandense do Arroz) for providing the rice seeds, for access to its facilities and for technical support.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Marcelo Menossi.
Raul Antonio Sperotto, Artur Teixeira de Araújo Junior and Janete Mariza Adamski contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.

299_2017_2234_MOESM1_ESM.jpg
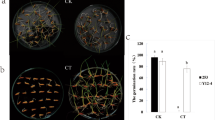
Fig. 1. Visual symptoms of rice plants from the cold-tolerant (CT) and cold-sensitive (CS) genotypes after 10 days under control (28 °C) or cold (10 °C) treatment, followed by 7 days of recovery at 28 °C. Bar in figure (A) indicates 7 cm. The experiment was performed twice with similar results (JPG 2062 KB)

299_2017_2234_MOESM2_ESM.jpg
Fig. 2. Analysis of genes expressed in leaves from two indica rice genotypes (cold-tolerant (CT) and cold-sensitive (CS)). (A) Scatter plot comparing the gene expression levels between the CT and the CS genotypes. (B) Genes identified by red dots have adjusted p-value lower than 0.000001 (calculated using EdgeR). FC: fold change; CPM: counts per million (JPG 2368 KB)
299_2017_2234_MOESM4_ESM.xlsx
Table 2. Differentially expressed genes revealed by RNAseq in leaves of rice plants exposed to cold treatment (10 °C) for 6 h. First panel: genes with higher expression in the cold-tolerant genotype (IRGA 959-1-2-2F-4-1-4-A). Second panel: genes with higher expression in the cold-sensitive genotype (IRGA 959-1-2-2F-4-1-4-D-1-CA-1) (XLSX 296 KB)
299_2017_2234_MOESM5_ESM.xlsx
Table 3. Meta-analysis of cold-responsive genes. All genes considered differentially expressed in this work (with higher expression in the cold-tolerant or in the cold-sensitive genotypes after plant exposure to 10 °C for 6 h) were used in searches for matching genes in tables of differentially expressed genes (in rice plants under cold treatment) reported in previous large-scale research papers. Details from each experiment are provided in the third spreadsheet. In the first two spreadsheets, genes that were reported as cold-responsive in at least one of the previous works are shown in red letters. Results obtained with rice genotypes considered cold-tolerant are shaded in blue, while results from cold-sensitive genotypes are shaded in yellow. Results obtained with rice Nipponbare are not shaded, because there is controversy about its sensitivity to cold. In the fourth spreadsheet, gene locations were compared with locations of QTLs previously identified as related to cold tolerance in rice plants during the vegetative stage. Genes with corresponding locations (“hits”) are marked with X in the corresponding columns and are shown in blue letters (XLSX 376 KB)
299_2017_2234_MOESM6_ESM.xlsx
Table 4. Descriptions of genes/proteins present in the response interactome of the cold-tolerant indica rice plants. Nodes with differentially expressed genes (higher expression in the cold-tolerant than in the sensitive plants) revealed by RNAseq in leaves of rice plants exposed to cold treatment (10 °C) for 6 h have fold change (FC Tolerant/Sensitive) shown in column D. The description (Hit) and gene ontology of each node was obtained from the Rice Genome Annotation Project (RGAP) database. The data of node degree and betweeneess centrality were obtained with a plugin network analyzer in the Cytoscape software. The metabolism categories were determined using the KEGG database (XLSX 70 KB)
Rights and permissions
About this article
Cite this article
Sperotto, R.A., de Araújo Junior, A.T., Adamski, J.M. et al. Deep RNAseq indicates protective mechanisms of cold-tolerant indica rice plants during early vegetative stage. Plant Cell Rep 37, 347–375 (2018). https://doi.org/10.1007/s00299-017-2234-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-017-2234-9