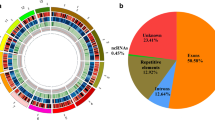
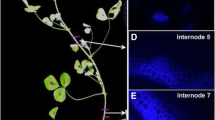
Abstract
A genome-wide inventory of proteins involved in cell wall synthesis and remodeling has been obtained by taking advantage of the recently released genome sequence of the ectomycorrhizal Tuber melanosporum black truffle. Genes that encode cell wall biosynthetic enzymes, enzymes involved in cell wall polysaccharide synthesis or modification, GPI-anchored proteins and other cell wall proteins were identified in the black truffle genome. As a second step, array data were validated and the symbiotic stage was chosen as the main focus. Quantitative RT-PCR experiments were performed on 29 selected genes to verify their expression during ectomycorrhizal formation. The results confirmed the array data, and this suggests that cell wall-related genes are required for morphogenetic transition from mycelium growth to the ectomycorrhizal branched hyphae. Labeling experiments were also performed on T. melanosporum mycelium and ectomycorrhizae to localize cell wall components.




Similar content being viewed by others
References
Aimanianda V, Bayry J, Bozza S, Kniemeyer O, Perruccio K, Elluru SR, Clavaud C, Paris S, Brakhage AA, Kaveri SV, Romani L, Latge JP (2009) Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460:1117–1121
Amicucci A, Balestrini R, Kohler A, Barbieri E, Saltarelli R, Faccio A, Roberson RW, Bonfante P, Stocchi V (2011) Hyphal and cytoskeleton polarization in Tuber melanosporum: a genomic and cellular analysis. Fungal Genet Biol 48:561–572
Balestrini R, Hahn MG, Bonfante P (1996) Location of cell-wall components in ectomycorrhizae of Corylus avellana and Tuber magnatum. Protoplasma 191:55–69
Balestrini R, Mainieri D, Soragni E, Garnero L, Rollino S, Viotti A, Ottonello S, Bonfante P (2000) Differential expression of chitin synthase III and IV mRNAs in ascomata of Tuber borchii Vittad. Fungal Genet Biol 31:219–232
Balestrini R, Bianciotto V, Bonfante P (2011) Mycorrhizae in: Huang, Li, Sumner (eds) Handbook of Soil Sciences, Volume I. Taylor and Francis Group, Boca Raton, pp 24/29–24/40
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795
Borgia PT, Iartchouk N, Riggle PJ, Winter KR, Koltin Y, Bulawa CE (1996) The chsB gene of Aspergillus nidulans is necessary for normal hyphal growth and development. Fungal Genet Biol 20:193–203
Borkovich KA, Alex LA, Yarden O et al (2004) Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev 68:1–108
Bowman SM, Free SJ (2006) The structure and synthesis of the fungal cell wall. BioEssays 28:799–808
Brendel V, Bucher P, Nourbakhsh IR, Blaisdell BE, Karlin S (1992). “Methods and algorithms for statistical analysis of protein sequences.” Proceedings of the National Academy of Sciences of the United States of America 89 (6) (March 15):2002–2006
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238
Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11:113–116
Choquer M, Boccara M, Goncalves IR, Soulié M-C, Vidal-Cros A (2004) Survey of the Botrytis cinerea chitin synthase multigenic family through the analysis of six euascomycetes genomes. Eur J Biochem 271:2153–2164
Coronado JE, Mneimneh S, Epstein SL, Qiu WG, Lipke PN (2007) Conserved processes and lineage-specific proteins in fungal cell wall evolution. Eukaryot Cell 6:2269–2277
Cserzo M, Eisenhaber F, Eisenhaber B, Simon I (2004) TM or not TM: transmembrane protein prediction with low false positive rate using DAS-TMfilter. Bioinformatics 20:136–137
de Groot PWJ, Brandt BW, Horiuchi H, Ramd AFJ, de Koster CG, Klis FM (2009) Comprehensive genomic analysis of cell wall genes in Aspergillus nidulans. Fungal Genet Biol 46:S72–S81
Denoeud F, Aury JM, Da Silva C, Noel B, Rogier O, Delledonne M, Morgante M, Valle G, Wincker P, Scarpelli C, Jaillon O, Artiguenave F (2008) Annotating genomes with massive-scale RNA sequencing. Genome Biol 9:R175
Duo-Chuan L (2006) Review of fungal chitinases. Mycopathologia 161:345–360
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Eisenhaber B, Bork P, Eisenhaber F (1999) Prediction of potential GPI-modification sites in proprotein sequences. J Mol Biol 292:741–758
Eisenhaber B, Schneider G, Wildpaner M, Eisenhaber F (2004) A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome-wide studies for Aspergillus nidulans, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae and Schizosaccharomyces pombe. J Mol Biol 337:243–253
Gruber S, Vaaje-Kolstad G, Matarese F, López-Mondéjar R, Kubicek CP, Seidl-Seiboth V (2011) Analysis of subgroup C of fungal chitinases containing chitin-binding and LysM modules in the mycoparasite Trichoderma atroviride. Glycobiology 21:122–133
Käll L, Krogh A, Sonnhammer ELL (2004) A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338:1027–1036
Karlsson M, Stenlid J (2008) Comparative evolutionary histories of the fungal chitinase gene family reveal non-random size expansions and contractions due to adaptive natural selection. Evol Bioinform 4:47–60
Kubicek C, Baker S, Gamauf C, Kenerley C, Druzhinina I (2008) Purifying selection and birth-and-death evolution in the class II hydrophobin gene families of the ascomycete Trichoderma/Hypocrea. BMC Evol Biol 8:4
Kulkarni RD, Kelkar HS, Dean RA (2003) An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. TIBS 28:118–121
Kurita T, Noda Y, Takagi T, Osumi M, Yoda K (2011) Kre6 protein essential for yeast cell wall β-1,6-glucan synthesis accumulates at sites of polarized growth. J Biol Chem 286:7429–7438
Laurent P, Voiblet C, Tagu D, de Carvalho D, Nehls U, De Bellis R, Balestrini R, Bauw G, Bonfante P, Martin F (1999) A novel class of ectomycorrhiza-regulated cell wall polypeptides in Pisolithus tinctorius. Mol Plant Microbe Interact 12:862–871
Lesage G, Bussey H (2006) Cell wall assembly in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 70:317–343
Martin F, Laurent P, de Carvalho D, Voiblet C, Balestrini R, Bonfante P, Tagu D (1999) Cell wall proteins of the ectomycorrhizal basidiomycete Pisolithus tinctorius: identification, function, and expression in symbiosis. Fungal Genet Biol 27:161–174
Martin F, Kohler A, Murat C, Balestrini R et al (2010) Périgord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature 464:1033–1038
Martin-Urdiroz M, Roncero MIG, Gonzalez-Reyes JA, Ruiz-Roldan C (2008) ChsVb, a Class VII chitin synthase involved in septation, is critical for pathogenicity in Fusarium oxysporum. Eukaryot Cell 7:112–121
Miozzi L, Balestrini R, Bolchi A, Novero M, Ottonello S, Bonfante P (2005) Phospholipase A2 up‐regulation during mycorrhiza formation in Tuber borchii. New Phytol 167:229–238
Montijn RC, Vink E, Muller WH, Verkleij AJ, Van Den Ende H, Henrissat B, Klis FM (1999) Localization of synthesis of β-1,6-glucan in Saccharomyces cerevisiae. J Bacteriol 181:7414–7420
Ooi HS, Kwo CY, Wildpaner M, Sirota FL, Eisenhaber B, Maurer-Stroh S, Wong WC, Schleiffer A, Eisenhaber F, Schneider G (2009) ANNIE: integrated de novo protein sequence annotation. Nucleic Acids Res 37:435–440
Pardo M, Monteoliva L, Vazquez P, Martinez R, Molero G, Nombela C, Gil C (2004) PST1 and ECM33 encode two yeast cell surface GPI proteins important for cell wall integrity. Microbiology 150:4157–4170
Pel HJ, De Winde JH, Archer DB et al (2007) Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 51388. Nat Biotech 25:221–231
Ragni E, Fontaine T, Gissi C, Latgè JP, Popolo L (2007) The Gas family of proteins of Saccharomyces cerevisiae: characterization and evolutionary analysis. Yeast 24:297–308
Ruiz-Herrera J, Ortiz-Castellanos L, Martínez A-I, León-Ramírez C, Sentandreu R (2008) Analysis of the proteins involved in the structure and synthesis of the cell wall of Ustilago maydis. Fungal Genet Biol 45:S71–S76
Seidl V (2008) Chitinases of filamentous fungi: a large group of diverse proteins with multiple physiological functions. Fungal Biol Rev 22:36–42
Seidl-Seiboth V, Gruber S, Sezerman U, Schwecke T, Albayrak A, Neuhof T, Von Döhren H, Baker SE, Kubicek CP (2011) Novel hydrophobins from Trichoderma define a new hydrophobin subclass: protein properties, evolution, regulation and processing. J Mol Evol 72:339–351
Shahinian S, Bussey H (2000) β-1,6-Glucan synthesis in Saccharomyces cerevisiae. Mol Microbiol 35:477–489
Soragni E, Bolchi A, Balestrini R, Gambaretto C, Percudani R, Bonfante P, Ottonello S (2001) A nutrient-regulated, dual localization phospholipase A2 in the symbiotic fungus Tuber borchii. EMBO J 20:5079–5090
Tagu D, De Bellis R, Balestrini R, de Vries OMH, Piccoli G, Stocchi V, Bonfante P, Martin F (2001) Immunolocalization of hydrophobin HYDPt-1 from the ectomycorrhizal basidiomycete Pisolithus tinctorius during colonization of Eucalyptus globulus roots. New Phytol 149:127–135
Takeshita N, Yamashita S, Ohta A, Horiuchi H (2006) Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor-like domain, perform compensatory functions that are essential for hyphal tip growth. Mol Microbiol 59:1380–1394
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thevissen K, Idkowiak-Baldys J, Im YJ, Takemoto J, François IE, Ferket KK, Aerts AM, Meert EM, Winderickx J, Roosen J, Cammue BP (2005) SKN1, a novel plant defensin-sensitivity gene in Saccharomyces cerevisiae, is implicated in sphingolipid biosynthesis. FEBS Lett 579:1973–1977
Treitschke S, Doehlemann G, Schuster M, Steinberg G (2010) The myosin motor domain of fungal chitin synthase V is dispensable for vesicle motility but required for virulence of the maize pathogen Ustilago maydis. Plant Cell 22:2476–2494
Tsigos I, Bouriotis V (1995) Purification and characterization of chitin deacetylase from Colletotrichum lindemuthianum. J Biol Chem 270:26286–26291
Tsigos I, Martinou A, Kafetzopoulos D, Bouriotis V (2000) Chitin deacetylases: new, versatile tools in biotechnology. Trends Biotech 18:305–312
Tusnády GE, Simon I (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850
van den Burg HA, Harrison SJ, Joosten MH, Vervoort J, de Wit PJ (2006) Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol Plant Microbe Interact 19:1420–1430
Weber I, Aßmann D, Thines E, Steinberg G (2006) Polar localizing class v myosin chitin synthases are essential during early plant infection in the plant pathogenic fungus Ustilago maydis. Plant Cell 18:225–242
Whiteford JR, Spanu PD (2002) Hydrophobins and the interactions between fungi and plants. Mol Plant Pathol 3:391–400
Zampieri E, Balestrini R, Kohler A, Abbà S, Martin F, Bonfante P (2011) The Perigord black truffle responds to cold temperature with an extensive reprogramming of its transcriptional activity. Fungal Genet Biol 48:585–591
Acknowledgments
The Tuber genome sequencing project is a collaborative effort involving the Génoscope-CEA and the Tuber Genome Consortium. The authors would like to thank Emmanuelle Morin and Stefano Ghignone for the bioinformatics support; Elisa Zampieri for the help during the preparation of the manuscript, Michele Buffalini, Paola Ceccaroli and Shwet Kamal for the annotation of some genes cited in the paper. This research was funded by PRIN2007; FS’s fellowship PhD was funded by Università di Torino.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by B. Cormack.
R. Balestrini and F. Sillo equally contributed to the paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
294_2012_374_MOESM4_ESM.pdf
Table S4. GPI-prediction by using the fungal-specific big-PI algorithm (Eisenhaber et al. 2004) and checking for a predicted signal peptide (Bendtsen et al. 2004). There is experimental evidence for alternative or secondary GPI-anchor attachment sites (Eisenhaber et al. 1999). As such, the predictor reports a main and a secondary site (1: and 2:), each one classified with a letter (P, predicted; S, twilight zone prediction; I, physical properties are not right, but profile matches; N, neither physical properties nor profile matches). The best hit is shown first and is either P or S: if the first one is P, the second one can be P, S or N, or if the first one is S it can be S, I or N. An additional step to check for potential transmembrane domains (TM) in the mature protein has been performed. (PDF 52 kb)
294_2012_374_MOESM5_ESM.xls
Table S5. Expression data for the GPI-anchored protein subset. The individual sheets contain the array and the Solexa/Illumina data as explained in the Supplemental file. (XLS 36 kb)
Rights and permissions
About this article
Cite this article
Balestrini, R., Sillo, F., Kohler, A. et al. Genome-wide analysis of cell wall-related genes in Tuber melanosporum . Curr Genet 58, 165–177 (2012). https://doi.org/10.1007/s00294-012-0374-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-012-0374-6