Abstract
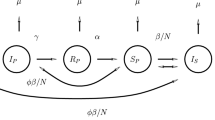
During an infection, HIV experiences strong selection by immune system T cells. Recent experimental work has shown that MHC escape mutations form an important pathway for HIV to avoid such selection. In this paper, we study a model of MHC escape mutation. The model is a predator–prey model with two prey, composed of two HIV variants, and one predator, the immune system CD8 cells. We assume that one HIV variant is visible to CD8 cells and one is not. The model takes the form of a system of stochastic differential equations. Motivated by well-known results concerning the short life-cycle of HIV intrahost, we assume that HIV population dynamics occur on a faster time scale then CD8 population dynamics. This separation of time scales allows us to analyze our model using an asymptotic approach. Using this model we study the impact of an MHC escape mutation on the population dynamics and genetic evolution of the intrahost HIV population. From the perspective of population dynamics, we show that the competition between the visible and invisible HIV variants can reach steady states in which either a single variant exists or in which coexistence occurs depending on the parameter regime. We show that in some parameter regimes the end state of the system is stochastic. From a genetics perspective, we study the impact of the population dynamics on the lineages of an HIV sample taken after an escape mutation occurs. We show that the lineages go through severe bottlenecks and that in certain parameter regimes the lineage distribution can be characterized by a Kingman coalescent. Our results depend on methods from diffusion theory and coalescent theory.
Similar content being viewed by others
References
Althaus C, de Boer R (2008) Dynamics of immune escape during HIV/SIV infection. PLOS Comput Biol 4(7): 1–10
Athreya K, Ney P (1972) Branching processes. Dover Publications, NY
Bhattacharya T et al (2007) Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science 315: 1583–1586
Crandal K (1999) The evolution of HIV. Johns Hopkins Press, Baltimore
DeFranco A, Locksley R, Robertson M (2007) Immunity: the immune response in infectious and inflammatory disease. New Science Press, USA
Delport W et al (2008) Frequent toggling between alternative amino acids is driven by selection in hiv-1. PLOS Pathog 4(12): 1–13
Durrett R (2002) Probability models for DNA sequence evolution. Springer, Berlin
Emini E (2002) The human immunodeficiency virus: biology, immunology, and therapy. Princeton University Press, NJ
Frahm N et al (2006) Control of HIV replication by CTL targeting subdominant epitopes. Nat Immunol 7(2): 173–178
Goulder P, Watkins D (2008) Impact of MHC class I diversity on immune control of HIV replication. Nat Rev 8: 619–629
Hartl D, Clark A (1997) Principles of population genetics, 3rd edn. Sinauer Associates, Sunderland
Ho D et al (1995) Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373: 123–126
Karatzas I, Shreve S (1991) Brownian motion and stochastic calculus. Springer, Berlin
Kouyos R et al (2006) Stochastic or deterministic: what is the effective population size of hiv-1. Trends Microbiol 14(12): 507–511
Kurtz T (1981) Approximation of population processes. In: 36, CBMS-NSF regional conference series in applied mathematics
Leigh-Brown A (1997) Analysis of HIV-1 env gene sequences reveals evidence for a low effective number in the viral population. PNAS 94: 1862–1865
Leslie A et al (2005) Transmission and accumulation of CTL escape variants drive negative association between HIV and HLA. J Exp Med 201(6): 891–902
Ngumbela K et al (2008) Targeting of a CD8 T cell env epitope presented by HLA-B*5802 is associated with markers of HIV disease progression and lack of selection pressure. AIDS Res Hum Retrovir 24(1): 72–82
Nowak M, May R (2000) Virus dynamics: mathematical principles of immunology and virology. Oxford University Press, Oxford
Perelson A (2002) Modeling viral and immune system dynamics. Nat Rev 2: 28–36
Rousseau C et al (2008) Hla class I-driven evolution of HIV type I subtype c proteome: immune escape and viral load. J Virol 82(13): 6434–6446
Rouzine I, Coffin J (1999) Linkage disequilibrium test implies a large effective population number for HIV in-vivo. PNAS 96: 10758–10763
Tavare S (2001) Ancestral inference in population genetics. St Flour probability summer school lecture notes. Springer, Berlin
Wakeley J, Sargsyan O (2009) The conditional ancestral selection graph with strong balancing selection. Theor Popul Biol 75: 355–364
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Leviyang, S. Analysis of a stochastic predator–prey model with applications to intrahost HIV genetic diversity. J. Math. Biol. 65, 1285–1336 (2012). https://doi.org/10.1007/s00285-011-0497-2
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-011-0497-2