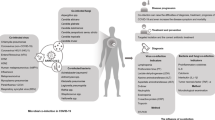
Abstract
The novel human coronavirus, Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), which results in the coronavirus disease 2019 (COVID-19), has caused a serious threat to global public health. Therefore, many studies are performed on the causes and prevalence of this disease and the possible co-occurrence of the infection with other viral and bacterial pathogens is investigated. Respiratory infections predispose patients to co‐infections and these lead to increased disease severity and mortality. Numerous types of antibiotics have been employed for the prevention and treatment of bacterial co‐infection and secondary bacterial infections in patients with a SARS-CoV-2 infection. Although antibiotics do not directly affect SARS‐CoV‐2, viral respiratory infections often result in bacterial pneumonia. It is possible that some patients die from bacterial co‐infection rather than virus itself. Therefore, bacterial co‐infection and secondary bacterial infection are considered critical risk factors for the severity and mortality rates of COVID‐19. In this review, we will summarize the bacterial co‐infection and secondary bacterial infection in some featured respiratory viral infections, especially COVID‐19.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Special Features of Coronavirus
The pandemic of severe acute respiratory syndrome coronavirus 2 raised the attention toward bacterial co-infection and its role in coronavirus disease 2019 (COVID-19) disease. The coronavirus belongs to the Coronaviridae virus family, infecting birds and mammals through respiratory infections [1]. This virus can cause common cold to more severe illnesses, such as SARS, MERS, and COVID-19 [2, 3]. The most prominent clinical symptoms of this disease are dry cough, shortness of breath, fever, and fluid accumulation in the interstitial space of the lungs and chest cavity [4, 5]. Respiratory viruses, such as coronaviruses and influenza viruses, can cause acute damage to lung epithelial cells, allowing other pathogens to infiltrate the affected area. [6, 7]. The history of COVID-19 infection is as follows: the first cases were reported in December 2019, from December 18 to 29, 2019, five patients were admitted with acute respiratory symptoms, one of whom died [2]. On January 2, 2020, a total of 41 patients with infectious symptoms were admitted to Wuhan Hospital in China and confirmed positive for COVID-19 by molecular tests [2]. Diabetes, hypertension, cardiovascular disease, cancer, lung disease, and other medical conditions were present in these patients [2]. It was assumed that these patients probably acquired nosocomial infections, which might be a reason for the severity of their disease [2]. Fever, cough, muscle, or body aches are common symptoms of coronavirus disease, while other symptoms include sputum, headache, bleeding, diarrhea, indigestion, and lymphopenia (decreased lymphocyte count) [8]. Lung ultrasound (LUS) is proven to be a valuable tool to detect specific findings in COVID-19 patients such as pneumonia [8]. Shortness of breath and muscle pain have also been reported in cases of systematic and severe implications [9, 10]. Abnormal features such as acute respiratory syndrome, acute heart failure, and fibrotic pulmonary lesions that lead to death have also been observed in these patients [11]. Sometimes, multiple ground glass opacity lesions have occasionally been observed in the sub-pleural regions of both lungs (accumulation of fluid in the space between the lungs and the chest cavity), leading to increased inflammation of this organ [12]. Some patients infected with COVID-19 also experienced gastrointestinal (GI) symptoms, such as diarrhea; however, a small percentage of patients infected with MERS or SARS viruses had experienced similar GI diseases [12]. Secondary bacterial infections play a critical role in the morbidity and mortality rates of patients initially falling ill with pulmonary viral diseases. This study aims to identify key characteristics of the microbial dysbiosis associated with patients infected with SARS-CoV-2. We have described not only the bacterial toxins, but also the direct bacterial invasion, which can be involved in severe COVID-19. Furthermore, this review discusses the interaction between SARS-CoV-2 and the host immune system.
Incidence of Microbial Diseases and Secondary Infections in Patients with COVID-19
Multiple ground glass opacity lesions have occasionally been observed in the sub-pleural regions of both lungs (fluid accumulation between the lungs and the chest cavity), leading to increased inflammation of this organ. The mechanism of action of viruses in the host body and the occurrence of symptoms were described in the preceding section. It can be noted that the proliferation of virus in lung tissue and its destructive effects on respiratory epithelial cells provide a suitable environment for the invasion and colonization of bacterial pathogens. [6]. So bacterial products may enter the patient’s bloodstream and cause more severe symptoms [13]. Bacterial and fungal secondary infections that co-occur with viral respiratory infections are attributed to mechanical ventilation used for the treatment of patients. Lung damage by COVID-19 and impaired immune response can provide a favorable environment for the proliferation and establishment of microorganisms in hospitalized patients. Studies show that secondary bacterial pneumonia is a potential risk factor for the severity and complications in patients with COVID-19 [14]. For example, secondary infections have been caused by Stenotrophomonas maltophilia, Klebsiella pneumonia (K. pneumonia), or Escherichia coli (E. coli) with serious complications, such as bacteremia, sepsis and nosocomial pneumonia in patients with SARS [15]. In addition, nosocomial infections and high mortality rate observed in influenza pandemics show that the entry of pathogenic populations such as Streptococcus pneumoniae, (S. pneumonia), Haemophilus influenzae (H. influenza), and Staphylococcus aureus (S. aureus) into the infected host is the causative factor [16]. Therefore, concerns about co-infection of SARS-CoV-2 with other known pathogenic viruses, bacteria, and fungi are discussed. Mechanical ventilation is required for the treatment of patients with acute respiratory symptoms and hypoxia, which can increase the risk of pneumonia. [17, 18]. Under these conditions, changes in oxygen levels, alveolar ventilation and deposition of inhaled particles, blood flow, and the density of inflammatory cells affect the microbial growth conditions in the lungs [19]. So far, the predominant microbial population observed in patients with severe COVID-19 infection includes Burkholderia cepacia complex (BCC), Staphylococcus epidermidis (S. epidermidis), Mycoplasma spp. (M. hominis, M. orale), S. pneumoniae, and H. influenzae [6, 14]. In addition, viral pathogens such as α herpesvirus 1, rhinovirus B, and human orthomyxovirus, as well as Aspergillus flavus and Candida fungi were isolated from these patients [18, 20]. Among all bacterial species, the most abundant is attributed to the Proteobacteria phylum [13]. According to Rawson et al., the overuse of broad-spectrum antibiotics creates an ideal environment for opportunistic bacterial colonization, secondary bacterial infections, and increased multidrug resistance (MDR) due to the destruction of the host’s normal flora, so that the prescription-based and self-administration of ineffective antimicrobial drugs during the COVID-19 pandemic have resulted in an increase in drug-resistant infections [21].
Determination of Microbial Population in Clinical Samples of Patients with COVID-19
Specifically, microbial cultures and laboratory tests on clinical samples have shown that opportunistic nosocomial pathogens such as nosocomial fungi and bacteria cause co-infection in patients with severe symptoms of COVID-19. Furthermore, the microbial population frequency in patients with severe and mild symptoms differs significantly. BCC was the most common respiratory pathogen found in the blood sample of patients causing nosocomial pneumonia (NP) in cystic fibrosis (CF) patients. [22, 23]. Moreover, the most important isolated pathogen from the nasal secretions of patients was S. epidermidis, which is reported to be a common skin bacterium and is known as an MDR agent [6]. The number of species of respiratory bacteria with moderate virulence, in particular, differed significantly from the pathogenic bacteria with high virulence. Notably, the number of respiratory microbial taxa detected (at the genus and species levels) was significantly higher in mild cases than in severe cases [6]. This is due to the widespread use of antimicrobial drugs during the treatment of patients with severe symptoms. Elimination and migration processes are two important determinants of the microbial population in healthy lungs, but in advanced lung diseases, the pathogenesis of the lung results from respiratory dysbiosis [19]. During the hyperventilation treatment process, the invasion of air-borne germs accelerates. Furthermore, the ability to remove microbes by bronchoconstriction is reduced by the disruption of mucus secretion. Therefore, this situation provides a nutrient-rich growth environment and, on the other hand, reduces the oxygen concentration in the presence of bacterial populations. Response to the inflammation and the damage to endothelial and epithelial lung tissue increase vascular permeability. This condition promotes the entry of bacteria into the alveolar region, resulting in growth and proliferation of specific lung pathogens [14]. Most of the microbiota in the body of patients with COVID-19 consisted of fifty most common species that are listed in Table 1. Among these species, the most typical respiratory pathogen is Acinetobacter baumannii (A. baumannii) followed by S. pneumoniae, P. aeruginosa, and S. aureus. Nosocomial pathogens mainly include fungi such as Candida albicans (C. albicans), Candida tropicalis (C. tropicalis), and bacteria including E. coli and Enterococcus faecalis (E. faecalis). The presence of these pathogens are mostly reported from China, France, Iran, and Brazil [14]. Coprobacillus and Delftia acidovorans were identified among opportunistic microorganisms, which are not usually pathogenic but are associated with catheter-related bacteremia and lung cavities formation [24, 25]. The predominant respiratory tract bacteria are identified in mild cases, including Veillonella, Neisseria, Streptococcus, and Prevotella, which are similar to the microbial flora of nose and oral cavity in healthy adult. Except for Veillonella, three other bacteria have been identified in patients with severe respiratory symptoms [6, 26, 27]. According to Sirivongrangson et al., 98.3% of COVID-19 patients have bacterial DNA in their blood serum. The predominant genera of Proteobacteria phylum—a major phylum of Gram-negative bacteria—are Sphingomonas, Bradyrhizobium (Bradyrhizobium enterica (B. enterica) were found in patients with colitis), Enhydrobacter, Phyllobacterium, Agrobacterium, Comamonas, Sediminibacterium, Acinetobacter, and Pseudomonas. COVID-19 may cause sepsis-like syndrome with a dominant bacterial population in sepsis, including Enhydrobacter, Bradyrhizobium, and Sphingomonas [13] (Table1). Sphingomonas paucimobilis is an opportunistic pathogen associated with nosocomial infections. B. enterica is found in patients with colitis and Enhydrobacter aerosaccus was isolated from patients with hemophagocytic lymphohistiocytosis (HLH) taking corticosteroids; therefore, the presence of HLH syndrome in patients with acute COVID-19 was taken into account. [28, 29].
Interaction of Bacterial Pathogens in Patients with COVID-19
Bacterial pathogens invading Covid-19 patients including A. baumannii, S. pneumoniae, P. aeruginosa, and S. aureus use their virulence factors in different ways, such as evasion of the host immune system, biofilm formation, induction of hemolysis, pneumolysin, phospholipases, iron absorption factors, stimulation of cytokines, adhesions, and the complementary resistance systems [30,31,32]. In pulmonary and catheter-related infections, biofilm formation in endotracheal tubes and mechanical ventilators is regarded as a bacterial interaction. As a result, a diverse microbial accumulation develops resistance to antimicrobial agents [33, 34]. The predominant biofilm-producing microorganisms in respiratory catheters are P. aeruginosa, S. aureus, and C. albicans. Species with the ability to form biofilms are much more resistant to antimicrobial and medicinal compounds [35]. For some of these microorganisms, biofilm production, expression of virulence factors, and antibiotic resistance are associated with oxygen restriction and the activity of macrophages [35]. Biofilm formation in S. aureus is described in association with the accumulation of activated macrophages with anti-inflammatory and pro-fibric properties. It is noteworthy that S. aureus biofilm exhibits the ability to polarize macrophages toward the alternatively activated (M2) macrophages, which are distinct from the classically activated (M1) macrophages. M1-macrophages are considered pro-inflammatory macrophages involved in host defense against bacterial infections, whereas M2-macrophages are responsible for anti-inflammation and bacterial persistence. Activation of M2-macrophages is usually associated with abundant expression and secretion of anti-inflammatory factors, such as Arginase-1, IL-10, and IL-12, and therefore, restrains a pro-inflammatory immune response [14]. During biofilm formation, bacterial species show resistance to the host immune response and inhibit phagocytosis by phagocytes and neutrophils [36]. Some pathogens, such as E. coli and E. faecalis, activate another important virulence factor called hemolysin, a protein that causes erythrocyte lysis and destruction of the cell membrane of lung tissue, causing pneumonia in patients with severe disease [37]. This bacterial virulence factor is produced when the level of lung surfactant decreases and as a result, it causes lung epithelial cell damage and induced the production of interleukin-8 (IL-8) [14, 37]. Another virulence factor is the pneumolysin toxin, which causes the formation of a pore in lung tissue. This protein is vital for virulence, in chronic bacteremia, and in the incidence of apoptosis; pneumolysin along with capsular polysaccharide and adhesion factors are involved in the development of inflammatory responses and the complement system [31]. Virulence factors such as phospholipase and Candida spp. biofilm formation play an important role in the development of a secondary infection in COVID-19 patients [38]. Phospholipases are very important in the development of lung disease because it causes changes in composition of cellular membrane, stimulates cytokines and chemokines, and involves in transport and exchange of respiratory gases in the blood [39]. In general, the success of bacteria in causing secondary infection in patients with COVID-19 is associated with an extensive set of virulence factors, including outer membrane proteins, secretory systems, surface adhesins, glycoconjugate, and iron absorption activities; therefore, they have an essential role in the development of antibiotic resistance during the infection [30]. In the metabolic processes of the tricarboxylic acid (TCA) cycle of biofilm-producing pathogens, the amount of dihydrolipoamide dehydrogenase (DLD) enzyme increases. The presence of DLD is essential for causing infection by pneumococcal species because it plays an essential role in the production of capsular polysaccharides and in the survival of S. pneumoniae in vivo [40]. Besides the enzymes associated in biofilm formation, other virulence factors such as adhesins, antibiotic-transporting proteases, toxins, and superantigens are identified during the exacerbation of secondary lung infection in patients with COVID-19. Herein, the sortase enzymes are an important virulence factor in Gram-positive bacteria that display adhesive proteins of bacteria into host cells [41]. In addition, virulence factors such as streptolysin S impair the function of phagocytes and promote the cytotoxic properties of epithelial cells [14]. Aside from the factors mentioned, various membrane transmitters, including protein secretion systems of types I, V, and IV, have been identified as being involved in the transfer of cytolysin-activating and adhesive proteins, which significantly increases the risk of infection in Covid-19-infected patients [14]. Dickson et al. point out that various lung diseases can alter the growth of localized microbiota, which increases the frequency of pathogenic bacteria populations and viral attacks in that area [19]. The most important contributing factors to the progression of respiratory diseases have been identified as neutrophil dysfunction, macrophage depletion, and excessive inflammation [42]. The evacuation of macrophages provides a suitable niche for secondary infection followed by deadly pneumonia caused by S. pneumoniae [42]. On the other hand, neutrophil dysfunction is caused by altered activity of angiotensin-converting enzyme 2 (ACE2); in pulmonary bacterial infections, this is caused by the invasion of pathogens, such as K. pneumoniae, H. influenzae, and S. aureus. When the level of active ACE2 changes, neutrophil infiltration increases and host defense capability and inflammatory responses intensify [43]. On the other hand, ACE2 converts angiotensin II to angiotensin‑(1‑7). After binding of SARS‑CoV‑2 virus to ACE2, the ACE2 levels decreases; therefore, the angiotensin II is not converted to angiotensin‑(1‑7) (1 and 3). Following the reduction in angiotensin-(1‑7) level, nitrogen radical production, and viral genome degradation, these processes will be terminated. In ACE2 deficiency conditions, the cellular signaling between angiotensin II and angiotensin‑(1‑7) will create an imbalance toward angiotensin II signaling. Angiotensin II increases the inflammation response by producing inflammatory cytokines and reactive oxygen species, which cause pneumonia and cell death [9]. The reactions seen in patients with a bacterial lung infection are similar to patients with COVID-19. It is noteworthy that people with chronic obstructive pulmonary disease (COPD) have complications, such as vascular permeability, platelet activation, and thrombosis, and expression of adhesion molecules, leukocyte utilization, and complement activation are also observed. There is also an increase in heme and heme oxygenase-1 (HO) in their blood, and the role of hypoxia-hemoglobinopathy and hyperferritinemia in exacerbating COVID-19 infection has been proven [44,45,46,47].
Interactions of Bacterial LPS Structure with SARS-CoV-2 Viral Proteins
COVID-19 protein, spike (S), binds to bacterial lipopolysaccharide and increases anti-inflammatory activity [48]. There is a direct link between high levels of LPS in the blood, the prevalence of metabolic syndrome, and the development of metabolic syndrome, all of which make people more susceptible to COVID-19 [48]. Low bacterial LPS mixed with S protein causes NF-B nuclear factors to activate more quickly, trigger cytokine responses in THP-1 monocytes [48], and increase blood mononuclear cells. Spike glycoprotein (S protein) is the most critical surface protein in coronaviruses, including SARS-CoV-2, which can interact with the ACE2 enzyme to facilitate virus entry into human respiratory epithelial cells [49] (Fig. 1). Acute respiratory distress syndrome (ARDS) is a general systemic inflammatory reaction found in many diseases, such as pneumonia, severe infections, and sepsis, in people with metabolic syndrome and COVID-19. Toll-like receptor 4 (TLR4) activation during ARDS by lipopolysaccharide stimulation causes the pro-inflammatory phase, which is characterized by the high release of cytokines, acute phase proteins, and reactive oxygen species [50]. The MST studies showed that the S protein of SARS-CoV-2 binds to the bacterial LPS. S protein also enhances the inflammatory response when LPS levels in THP-1 monocytes and PBMCs are low in human blood [48]. With increasing levels of LPS and protein S in the plasma of patients, it has been observed that the rate of AP-1/NF-κB (nuclear factor kappa-light-chain enhancer of activated B cells) increases significantly; this can be seen even when both factors are present in low quantity. On the other hand, low LPS and S protein levels significantly increase tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6) as well as the IL-8 and interferon beta (IFN-β) responses [48]. Studies in the lab have demonstrated that S protein enhances monocyte responsiveness to LPS. In addition, the increase in cytokine levels is directly related to activation of NF-κB [51]. Thus, activation and increase of NF-κB level by increasing the amount of protein S and LPS significantly increases the inflammatory response and produces a long-term response [52]. As a result of these events, protein S and LPS are broken down, and high amounts of LPS are accumulated which is proven by fluorescein-labeled LPS (LPS-FITC) [48].
a Schematic diagram of the SARS-CoV-2 coronavirus particle. Four structure proteins contain the envelope protein (E), nucleocapsid protein (N), spike protein (S), and membrane protein (M). SARS-CoV-2 virus enters the cell through the S protein on the surface of SARS-CoV-2 by means of binding with its receptor ACE2. Proteins (S proteins) affected with genetic alterations means that the SARS-CoV-2 spike protein mediates target recognition, cellular entry, and ultimately the viral infection that leads to various levels of COVID-19 severities. Positive evolutionary selection of mutations within the spike protein has led to the genesis of new SARS-CoV-2 variants with greatly enhanced overall fitness. b Schematic image of SARS-CoV-2 virus showing the Spike (S) protein which is heavily glycosylated (O-glycosylation and N-glycosylation) with numerous fucose, mannose, and sialyl residues. These adhesion have the potential to bind to C-type lectin receptors (CLR), mannose receptor (MR), dendritic cell-specific intracellular adhesion molecule-3-grabbing non-integrin (DC-SIGN), homologue dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin related (L-SIGN), macrophage galactose-type lectin (MGL), toll-like receptors (TLR), and glucose regulated
Endotoxemia and Circulating Bacterium in Acute COVID-19 Patients
While the respiratory tract is the leading site of COVID-19 infection, the gastrointestinal tract of many patients is also involved and causes symptoms, such as diarrhea and bloating [53]. In the stool samples obtained from COVID-19 patients, the presence of bacterial agents in the gastrointestinal tract may make the symptoms of acute COVID-19 disease more difficult to manage which is in accordance with the findings of the intestinal ileum’s enterocytes and the colon’s expressed ACE2 receptor [6]. Endotoxin is a component of the cell wall of the Gram-negative bacteria, which is considered as a leading cause of death in patients with COVID-19 due to sepsis shock and is regarded as the primary key for stimulation and the attack of cytokines [54]. Thus, bacterial toxins and direct bacterial invasion may play a role in worsening the complications of acute COVID-19 disease [13]. Microbial effects in patients can be monitored using serum factor (1 → 3) β-D-glucan (BG) which is a marker for intestinal dysfunction and endotoxin activity assay (EAA). Patients with COVID-19 and bacterial infections have been specifically reported to have elevated serum levels of BG and EAA [13]. Generally, in this group of patients, the sputum culture and endotracheal suction are positive for COVID-19 [13]. Moreover, for the treatment of patients with high EAA, more mechanical ventilation is needed compared to patients with low EAA. BG is a major structural polysaccharide of the cell wall of most fungi, including Candida spp., which is used as a biomarker to detect intestinal permeability in human sepsis. Elevated serum BG levels in patients with COVID-19 and exposure to antibiotics may be a sign of overgrowth of Candida in their gastrointestinal tract. On the other hand, the presence of BG can trigger pro-inflammatory responses through Dectin-1 signaling [55, 56]. The activity of endotoxins in viral infections is of particular importance, as various studies show that the presence of LPS in patients with Influenza virus infection activates pulmonary inflammatory responses. This leads to secondary bacterial pneumonia because LPS binds to TLR4 receptors and activates transcription factors such as activating protein 1 (AP-1), NF-κB, interferon regulatory factor 3 (IRF3), myeloid differentiation factor 88 (MyD88), and TIR-domain-containing adapter-inducing interferon-β (TRIF)-dependent pathway. This leads to the induction of pro-inflammatory cytokines and function of interferon [57]. High levels of LPS and 16S rRNA bacterial gene product are found in patients with severe COVID-19 compared to healthy individuals. The number and diversity of bacterial populations are important factors in the intensification of COVID-19. Given that the high levels of pro-inflammatory cytokines, including IL-6, IL-1β, IP10, and MCP-1, in patients with COVID-19, it can be stated that bacterial products are another contributing factor to the cytokines storm along with viral agents [58]. Although the source of toxins and bacterial DNA in the blood of patients with COVID-19 pneumonia is unknown, viremia may lead to capillary leak syndrome similar to bacterial sepsis, which causes interstitial edema and induces lung dysfunction and intestinal barrier. This could be the pathway that allows bacterial toxins and live bacteria enter the bloodstream, followed by the release of toxins that cause pro-inflammatory responses [13]. COVID-19 also targets the gastrointestinal tract (GI) in addition to the respiratory tract. Similarly to the respiratory system, gastrointestinal cells express ACE2 and TMPRSS2, which are required for viral particle integration with host receptor cells [59, 60]. Therefore, losing the intestinal defense barrier may be one mechanism that causes the presence and release of bacterial toxins and bacterial DNA in the blood of patients with acute COVID-19 (Fig. 2). SARS-CoV-2 infection of enterocytes causes inflammatory response of gastrointestinal tract which results in alteration of the intestinal microenvironment, including epithelial hyper permeability, attenuated local immune system, and dysbiosis of the microbiome [13]. Biopsies from severe cases have revealed the presence of COVID-19 throughout the GI tract from esophagus to colon [61]. Thus, loss of gut barrier function might be one of the mechanisms that contribute to the presence of bacterial toxin and bacterial DNA in the blood of patients with severe COVID-19 [61]. SARS-CoV-2 causes novel multisystem inflammatory syndrome (MIS) with symptoms associated with persistent fever and severe inflammation. The clinical similarity of MIS toxic shock syndrome (TSS) due to the activity of a bacterial superantigen (SAg) suggests that SARS-CoV-2 may have a SAg-like motif that plays a role in the severity of symptoms or in the stimulation of the autoimmune-like mechanism causes excessive inflammation [62, 63]. Comparing with the bacterial toxins, a motif in S protein showed that SARS-CoV-2 has a very similar structural sequence to a part of the bacterial SAg of Staphylococcal enterotoxin type B (SEB). The position of the SAg-like motif in the S protein of SARS-CoV-2 is of high importance.
Pathogenesis of endotoxemia in Covid-19 patient. At early stage, SARS-CoV-2 primarily infects type 2 pneumocytes in the lungs and causes pneumonia which can progress to acute respiratory distress syndrome (ARDS) and induces susceptibility of secondary bacterial infection by impairing the pulmonary immune response. The perturbations of the intestinal microenvironment allow the translocation of pathogenic bacteria from the gut lumen to the bloodstream. This figure was generated using the BioRender (available at https://biorender.com/)
Each HCoV (human coronavirus) promoter comprises two subunits, S1 and S2, each of which has a different role in causing a viral infection. The S1 subunit contains the Receptor-binding domain that binds to its specific receptors (ACE2 in SARS-CoV-2, SARS-CoV, and HCov-NL63) on the host cell [62, 64]. The S2 subunit contains the fusion peptide, which is essential for the virus to enter the host cell [65, 66]. The SAg-like motif is in the C-terminal domain of the S1 subunit at the border with S2 [62]. Membrane fusion requires two consecutive cleavages by host cell proteases, one in the S1/S2 interface (peptide bond R685-S686) and the other in the S2́ site (R815-816), thus, the SAg-like motif site overlaps with the S1/S2 cleavage site at the S-glycoprotein level [65, 67, 68]. Another unique feature of the SARS-CoV-2 structure is an insertion called the 681 PRRA 685 in close proximity to the SAg-like motif, adjacent to the R685-S686 cleavage area [62]. SARS-CoV-2 is the only βCov strain that contains this insertion [62]. The PRRA insertion sequence is very flexible, forming a highly active part of 681PRRAR685 together with the lateral arginine sequences [62]. This section plays a crucial role in identifying and binding host cell proteases, including transmembrane protease serine 2 (TMPRSS2) and furin, the cleavage of which is essential for the initiation of S protein activity [69, 70] (Fig. 3).
Immune mechanism affected by Covid-19 in different stages. In viremia: The virus spreads through the body and there is excessive activation of immune cells, such as IFN γ, IL-2 and TNFα, triggering cytokine storms and immune impairment. In acute phase: Characterization by the appearance of Covid-19 symptoms presents a profile of immune cells, such as Tim3, PD1, TIGIT, and NKG2A. In this period, the presence of CD14 + and CD16 + hyper inflammatory monocytes, with a high production capacity of TNFα, IL-1β and IL-6, are seen, which will migrate to the lungs. In convalescence: The individual can evolve in two opposite directions, recovery or clinical worsening/death. In recovery, cells of lymphoid origin recover their effector function and lose markers of exhaustion, while IgG levels improve. This figure was generated using the BioRender (available at https://biorender.com/)
Antibody Response Against SARS-CoV-2
The 681PRRAR685 polybasic site is thought to be involved in critical interactions with host cell proteins so that it can act as a target for S protein-neutralizing antibodies (Abs) SARS-CoV-2 (S-neutralizing antibodies) [62]. Given the sequence and structural similarity between the PRRA insertion sequence of the SAg-like motif and the bacterial enterotoxin SEB, it is hypothesized that the monoclonal anti-SEB antibody (mAbs), which can bind to the SAg-like viral motif, in particular the 681PRRAR685 fragment, is pre-produced and may block access to the S1/S2 cleavage site [62]. In the immune response to pathogens, factor 6D3 (PRDM1/Blimp-1 antibody) is shown to inhibit virus entry into the host cell. As a result, it is seen that the 6D3-binding section and the aforementioned protease region overlap. Consequently, the problem is that 6D3 might prevent the virus from entering. On the other hand, SEB-specific mAb 6D3 is demonstrated to have a strong tendency to bind to the S1/S2 site [62]. 6D3 may also target the RBD or other non-overlapping components in combination with other Abs neutralizers, which increases the effect of mAbs by inhibiting the entry of SARS-CoV-2 into the cell [62, 71]. In addition to the 6D3, 4A8 anti SARS-CoV-2 spike (NTD) antibody can also be attached to similar sections, blocking the S1/S2 Section [62]. The proximity of the viral SAg-like site to the S1/S2 proteolytic cleavage site (peptide bond 685R-S686) suggests that an anti-SEB mAb that cross-react with SARS-CoV-2 may have the potential to block the incision area of the virus. It also modulates cytokine storms and subsequent pro-inflammatory reactions [72]. Three SEB-related mAbs called 14G8, 6D3, and 20B1 have been proposed as the effective SAg toxin B activity inhibitors in animal models. These antibodies bind to different parts of the SEB. Of these antibodies, only 6D3 is structurally and persistently similar to the SARS-CoV-2 S SAg-like motif [73]. Computational analysis studies by Cheng et al. indicate that 6D3 antibody has a stronger tendency to bind to the SAg polybasic site than TMPRSS2 and furin [62, 71]. Based on the results of immunological studies on SARS-CoV-2, it has been shown that the 6D3 antibody can prevent COVID-19 viral infection (Fig. 4).
Sequence alignment of SARS-CoV-2 and multiple SARS-related strains near the insertion of PRRA. Sequence alignment of SARS-CoV-2 is near the S1/S2 cleavage site against multiple bat and pangolin SARS-related strains. Note that the polybasic insert PRRA of SARS-CoV-2 S is not found in closely related SARS-like CoVs, but exists in MERS and HCoVs HKU1 and OC43 (Red fonts). The furin-like cleavage site is indicated by the blue-shaded box (Color figure online)
Conclusion
Distinct differences in microbial composition are observed between patients infected with COVID-19 having low or severe virulence in their respiratory and gastrointestinal systems. One reason for these differences is the use of antibiotics and, on the other hand, the development of antibiotic resistance by some bacterial species. Bacterial populations of BCC and S. epidermidis and Mycoplasma spp. have been identified as opportunistic respiratory pathogens in patients with acute COVID-19 and patients with long-term admission to ICU intensive care unit. These bacteria have even been mentioned as significant cause of death in these patients. On the other hand, the genera Veillonella, Neisseria, Streptococcus, and Prevotella in a mild form are the dominant bacteria in the respiratory tract of patients [6]. The results of various studies show that the exposure to COVID-19 reduces microbial diversity but increases the pathogen population [14]. Therefore, the abundant respiratory pathogens are the bacteria including A. baumannii and S. pneumonia bacteria and fungi, such as C. albicans and C. tropicalis. The presence of C. albicans together with pathogenic bacteria in a synergistic interaction helps the fungal species readily absorb iron and contribute to further pathogenesis [14]. As a result, the presence of nosocomial opportunistic respiratory pathogens during the colonization process in the host body expresses and secretes a wide range of virulence factors, leading to the secretion of cytokines and the development of an inflammatory response [14]. In the early stages of SARS-CoV-2 infection, mainly type 2 pneumocytes become infected in the lungs, causing pneumonia, which can progress to ARDS and increase the risk of secondary bacterial infections with decreased lung immune response. The virus can enter the bloodstream and cause viremia, affecting various organs in the body, including the intestines, with high ACE2 expression. The SARS-CoV-2 infection causes an inflammatory response in the gastrointestinal tract and changes in the intestinal microenvironment. These changes include epithelial hyperpermeability, localized immunosuppression, and microbiome dysbiosis. These disorders allow pathogenic bacteria to enter the bloodstream from the intestinal lumen. Therefore, the sources of endotoxin isolated from COVID-19 patients are known to be the pathogenic bacteria in the intestinal tract causing secondary bacterial infections in the respiratory system. Immunological studies in patients with COVID-19 have shown that 6D3 antibody can block the S protein protease cleavage site of SARS-CoV-2 and prevent virus entry. On the other hand, virus S protein is structurally similar to SAg-like SEB. Therefore, the 6D3 antibody has the dual potential to block SARS-CoV-2 infection as well as SAg associated with T-cell activity and pro-inflammatory responses.
References
Abolghasemi H, Bashash D, Jafari R, Naseri P, Farzanehpour M, Bolandian M et al (2021) A comparative study of laboratory findings in PCR-positive and PCR-negative COVID-19 hospitalized patients. Irish J Med Sci 191:1–8
Rothan HA, Byrareddy SN (2020) The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun 109:102433
Akbariqomi M, Hosseini MS, Rashidiani J, Sedighian H, Biganeh H, Heidari R et al (2020) Clinical characteristics and outcome of hospitalized COVID-19 patients with diabetes: a single-center, retrospective study in Iran. Diabetes Res Clin Pract 169:108467
Sohrabi C, Alsafi Z, O’Neill N, Khan M, Kerwan A, Al-Jabir A et al (2020) World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19). Int J Surg (London, England) 76:71–76
Fallah A, Sedighian H, Behzadi E, Havaei SA, Kachuei R, Fooladi AAI (2022) The role of serum circulating microbial toxins in severity and cytokine storm of COVID positive patients. Microb Pathog 174:105888
Zhong H, Wang Y, Shi Z, Zhang L, Ren H, He W et al (2021) Characterization of respiratory microbial dysbiosis in hospitalized COVID-19 patients. Cell Discov 7(1):23
de Wit E, van Doremalen N, Falzarano D, Munster VJ (2016) SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 14(8):523–534
Liu F, Xu A, Zhang Y, Xuan W, Yan T, Pan K et al (2020) Patients of COVID-19 may benefit from sustained lopinavir-combined regimen and the increase of eosinophil may predict the outcome of COVID-19 progression. Int J Infect Dis 95:183–191
Hedayati MA (2020) Ambient temperature interferes with COVID-19. Int J Prev Med 1:109
Hedayati MA (2021) Viral respiratory coinfections enhance the spread of SARS-CoV-2. Infect Dis Clin Pract (Baltimore, Md) 29(6):e488
Xu J, Zhao S, Teng T, Abdalla AE, Zhu W, Xie L et al (2020) Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses 12(2):244
Lei J, Li J, Li X, Qi XJR (2020) CT imaging of the 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology 295(1):18
Sirivongrangson P, Kulvichit W, Payungporn S, Pisitkun T, Chindamporn A, Peerapornratana S et al (2020) Endotoxemia and circulating bacteriome in severe COVID-19 patients. Intensiv Care Med Exp 8(1):1–15
de Carvalho FM, Lemos LN, Ciapina LP, Moreira RG, Gerber A, Guimarães APC et al (2020) Prevalence of bacterial pathogens and potential role in COVID-19 severity in patients admitted to intensive care units in Brazil. Medrxiv. https://doi.org/10.1101/2020.12.22.20248501
Peiris JSM, Chu C-M, Cheng VC-C, Chan K, Hung I, Poon LL et al (2003) Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361(9371):1767–1772
Morris DE, Cleary DW, Clarke SCJ (2017) Secondary bacterial infections associated with influenza pandemics. Front Microbiol 8:1041
Meng L, Qiu H, Wan L, Ai Y, Xue Z, Guo Q et al (2020) Intubation and ventilation amid the COVID-19 outbreak: Wuhan’s experience. Anesthesiology 132(6):1317–1332
Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z et al (2020) Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet (London, England) 395(10229):1054–1062
Dickson RP, Martinez FJ, Huffnagle GB (2014) The role of the microbiome in exacerbations of chronic lung diseases. Lancet (London, England) 384(9944):691–702
Kim D, Quinn J, Pinsky B, Shah NH, Brown IJJ (2020) Rates of co-infection between SARS-CoV-2 and other respiratory pathogens. JAMA 323(20):2085–2086
Rawson TM, Moore LS, Zhu N, Ranganathan N, Skolimowska K, Gilchrist M et al (2020) Bacterial and fungal coinfection in individuals with coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis 71(9):2459–2468
Lee JY, Monk IR, da Silva AG, Seemann T, Chua KY, Kearns A et al (2018) Global spread of three multidrug-resistant lineages of Staphylococcus epidermidis. Nat Microbiol 3(10):1175–1185
Miragaia M, Couto I, Pereira SF, Kristinsson KG, Westh H, Jarløv JO et al (2002) Molecular characterization of methicillin-resistant Staphylococcus epidermidis clones: evidence of geographic dissemination. J Clin Microbiol 40(2):430–438
Yildiz H, Sünnetçioğlu A, Ekin S, Baran Aİ, Özgökçe M, Aşker S et al (2019) Delftia acidovorans pneumonia with lung cavities formation. Colomb Med (Cali) 50(3):215–221
Patel D, Iqbal AM, Mubarik A, Vassa N, Godil R, Saad M et al (2019) Delftia acidovorans: a rare cause of septic pulmonary embolism from catheter-related infection: case report and literature review. Med Case Rep 27:100835
Man WH, de Steenhuijsen Piters WA, Bogaert DJNRM (2017) The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Mocrobiol 15(5):259–270
Huffnagle G, Dickson R, Lukacs NW (2017) The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol 10(2):299–306
Mishra D, Satpathy G, Wig N, Fazal F, Ahmed NH, Panda SKJJoi, et al (2020) Evaluation of 16S rRNA broad range PCR assay for microbial detection in serum specimens in sepsis patients. J Infect Public Health 13(7):998–1002
Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ (2020) COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395(10229):1033–1034
Harding CM, Hennon SW, Feldman MF (2018) Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol 16(2):91–102
Kadioglu A, Weiser JN, Paton JC, Andrew PWJNRM (2008) The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol 6(4):288–301
Bien J, Sokolova O, Bozko P (2011) Characterization of virulence factors of Staphylococcus aureus: novel function of known virulence factors that are implicated in activation of airway epithelial proinflammatory response. J Pathog 2011:601905
Adair CG, Gorman SP, Feron BM, Byers LM, Jones DS, Goldsmith CE et al (1999) Implications of endotracheal tube biofilm for ventilator-associated pneumonia. Intensive Care Med 25(10):1072–1076
Wolcott R, Costerton JW, Raoult D, Cutler SJ (2013) The polymicrobial nature of biofilm infection. Clin Microbiol Infect 19(2):107–112
Hanke ML, Kielian T (2012) Deciphering mechanisms of staphylococcal biofilm evasion of host immunity. Front Cell Infect Microbiol 2:62
Hirschfeld J (2014) Dynamic interactions of neutrophils and biofilms. J Oral Microbiol 6:26102
Doran KS, Chang JC, Benoit VM, Eckmann L, Nizet V (2002) Group B streptococcal beta-hemolysin/cytolysin promotes invasion of human lung epithelial cells and the release of interleukin-8. J Infect Dis 185(2):196–203
Deorukhkar SC, Saini S, Mathew S (2014) Virulence factors contributing to pathogenicity of Candida tropicalis and Its antifungal susceptibility profile. Int J Microbiol 2014:456878
Hurley BP, McCormick BA (2008) Multiple roles of phospholipase A2 during lung infection and inflammation. Infect Immun 76(6):2259–2272
Smith AW, Roche H, Trombe MC, Briles DE, Håkansson A (2002) Characterization of the dihydrolipoamide dehydrogenase from Streptococcus pneumoniae and its role in pneumococcal infection. Mol Microbiol 44(2):431–448
Datta V, Myskowski SM, Kwinn LA, Chiem DN, Varki N, Kansal RG et al (2005) Mutational analysis of the group A streptococcal operon encoding streptolysin S and its virulence role in invasive infection. Mol Microbiol 56(3):681–695
Ghoneim HE, Thomas PG, McCullers JA (2013) Depletion of alveolar macrophages during influenza infection facilitates bacterial superinfections. J Immunol (Baltimore) 191(3):1250–1259
Sodhi CP, Nguyen J, Yamaguchi Y, Werts AD, Lu P, Ladd MR et al (2019) A dynamic variation of pulmonary ACE2 is required to modulate neutrophilic inflammation in response to Pseudomonas aeruginosa lung infection in mice. J Immunol 203(11):3000–3012
Aggarwal S, Ahmad I, Lam A, Carlisle MA, Li C, Wells JM et al (2018) Heme scavenging reduces pulmonary endoplasmic reticulum stress, fibrosis, and emphysema. JCI Insight. https://doi.org/10.1172/jci.insight.120694
Raval M, C, J Lee PJ, (2010) Heme oxygenase-1 in lung disease. Curr Drug Target 11(12):1532–1540
Su WL, Lin CP, Hang HC, Wu PS, Cheng CF, Chao YC (2021) Desaturation and heme elevation during COVID-19 infection: a potential prognostic factor of heme oxygenase-1. J Microbiol Immunol Infect 54(1):113–116
Wagener F, Pickkers P, Peterson SJ, Immenschuh S, Abraham NG (2020) Targeting the heme-heme oxygenase system to prevent severe complications following COVID-19 infections. Antioxidants (Basel, Switzerland) 9(6):540
Petruk G, Puthia M, Petrlova J, Samsudin F, Strömdahl A-C, Cerps S et al (2020) SARS-CoV-2 Spike protein binds to bacterial lipopolysaccharide and boosts proinflammatory activity. J Mol Cell Biol 12(12):916–932
Wan Y, Shang J, Graham R, Baric RS, Li F (2020) Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. https://doi.org/10.1128/JVI.00127-20
Rittirsch D, Flierl MA, Ward PA (2008) Harmful molecular mechanisms in sepsis. Nat Rev 8(10):776–787
Liu X, Yin S, Chen Y, Wu Y, Zheng W, Dong H et al (2018) LPS-induced proinflammatory cytokine expression in human airway epithelial cells and macrophages via NF-κB, STAT3 or AP-1 activation. Mol Med Rep 17(4):5484–5491
Puthia M, Butrym M, Petrlova J, Strömdahl A-C, Andersson MÅ, Kjellström S et al (2020) A dual-action peptide-containing hydrogel targets wound infection and inflammation. Sci Transl Med. https://doi.org/10.1126/scitranslmed.aax6601
Luo S, Zhang X, Xu HJCG (2020) Don’t overlook digestive symptoms in patients with 2019 novel coronavirus disease (COVID-19). Clin Gastroenterol Hepatol 18(7):1636
Opal S (2007) The host response to endotoxin, antilipopolysaccharide strategies, and the management of severe sepsis. Int J Med Microbiol 297(5):365–377
Leelahavanichkul A, Worasilchai N, Wannalerdsakun S, Jutivorakool K, Somparn P, Issara-Amphorn J et al (2016) Gastrointestinal leakage detected by serum (1→ 3)-β-D-glucan in mouse models and a pilot study in patients with sepsis. Shock 46(5):506–518
Netea MG, Joosten LA, Van Der Meer JW, Kullberg B-J, Van De Veerdonk FL (2015) Immune defence against Candida fungal infections. Nat Rev Immun 15(10):630–642
Koch R, Diavatopoulos D, Ferwerda G, Pickkers P, de Jonge M, Kox M (2018) The endotoxin-induced pulmonary inflammatory response is enhanced during the acute phase of influenza infection. Intensiv Care Med Exp 6(1):1–10
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y et al (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395(10223):497–506
Jin X, Lian J-S, Hu J-H, Gao J, Zheng L, Zhang Y-M et al (2020) Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut 69(6):1002–1009
Lin L, Jiang X, Zhang Z, Huang S, Zhang Z, Fang Z et al (2020) Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut 69(6):997–1001
Wang Q, Boshoff HI, Harrison JR, Ray PC, Green SR, Wyatt PG et al (2020) PE/PPE proteins mediate nutrient transport across the outer membrane of Mycobacterium tuberculosis. Science 367(6482):1147–1151
Cheng MH, Zhang S, Porritt RA, Noval Rivas M, Paschold L, Willscher E et al (2020) Superantigenic character of an insert unique to SARS-CoV-2 spike supported by skewed TCR repertoire in patients with hyperinflammation. Proc Natl Acad Sci 117(41):25254
Rivas MN, Porritt RA, Cheng MH, Bahar I, Arditi M (2021) COVID-19–associated multisystem inflammatory syndrome in children (MIS-C): a novel disease that mimics toxic shock syndrome—the superantigen hypothesis. J Allergy Clin immunol 147(1):57–59
Benton DJ, Wrobel AG, Xu P, Roustan C, Martin SR, Rosenthal PB et al (2020) Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 588(7837):327–330
Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E (2020) The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir Res 176:104742
Cui J, Li F, Shi ZL (2019) Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17(3):181–192
Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q (2020) Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (New York, NY) 367(6485):1444–1448
Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I et al (2020) Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci 117(13):7001–7003
Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D (2020) Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181(2):281–92.e6
Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A et al (2020) Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci 117(21):11727–11734
Min L, Sun QJ (2021) Antibodies and vaccines target RBD of SARS-CoV-2. Front Mol Biosci 8:247
Krakauer TJT (2019) Staphylococcal superantigens: pyrogenic toxins induce toxic shock. Toxin 11(3):178
Dutta K, Varshney AK, Franklin MC, Goger M, Wang X, Fries BC (2015) Mechanisms mediating enhanced neutralization efficacy of staphylococcal enterotoxin B by combinations of monoclonal antibodies. J Biol Chem 290(11):6715–6730
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or non-profit sectors.
Author information
Authors and Affiliations
Contributions
PF, HS, and AI wrote the manuscript with support from all authors. PF draw the figures. EB, HS, and RK revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fazel, P., Sedighian, H., Behzadi, E. et al. Interaction Between SARS-CoV-2 and Pathogenic Bacteria. Curr Microbiol 80, 223 (2023). https://doi.org/10.1007/s00284-023-03315-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03315-y