Abstract
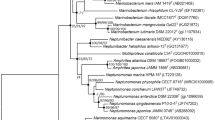
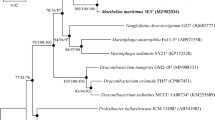
A Gram-stain-positive, yellow, aerobic, slender rod-shaped bacterial strain, designated KN1116T, was isolated from a deep-sea seamount. Phylogenetic analysis based on 16S rRNA gene sequence showed that strain KN1116T was related to the genus Chryseoglobus and had highest 16S rRNA gene sequence identity with Chryseoglobus frigidaquae CW1T (98.5%). The predominant cellular fatty acids were anteiso-C15:0 and iso-C16:0. The quinone system for strain KN1116T comprised menaquinone MK-12, MK-11, MK-10 and MK-13. The polar lipid profile contained diphosphatidylglycerol, phosphatidylglycerol, six unknown glycolipids, two unidentified phospholipids and one unknown polar lipid. The cell-wall peptidoglycan of strain KN1116T was of the type B1β, containing 2,4-diaminobutyric acid as the diamino acid. Genome sequencing revealed the strain KN1116T has a genome size of 2.7 Mbp and a G+C content of 69.4 mol%. Based on phenotypic, chemotaxonomic, phylogenetic and genomic data, strain KN1116T represents a novel species of a novel genus of the family Microbacteriaceae, for which the name Marinisubtilis pacificus gen. nov., sp. nov. is proposed. The type strain of Marinisubtilis pacificus is KN1116T (=CGMCC 1.17143T =KCTC 49299T).


Similar content being viewed by others
References
Park YH, Suzuki K, Yim DG, Lee KC, Kim E et al (1993) Suprageneric classification of peptidoglycan group B actinomycetes by nucleotide sequencing of 5S ribosomal RNA. Antonie Van Leeuwenhoek 64:307–313
Stackebrandt E, Rainey FA, Ward-Rainey NL (1997) Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol 47:479–491
Zhi XY, Li WJ, Stackebrandt E (2009) An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int J Syst Evol Microbiol 59:589–608
Schleifer KH, Kandler O (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36:407–477
Evtushenko LI, Takeuchi M (2006) The family Microbacteriaceae. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E et al (eds) The prokaryotes, vol 3, 3rd edn. Springer, New York, pp 1020–1098
Evtushenko LI (2012) Family XI Microbacteriaceae. In: Goodfellow M, Kämpfer P, Busse HJ, Trujillo ME, Suzuki K et al (eds) Bergey’s manual of systematic bacteriology, vol 5, 2nd edn. Springer, New York, pp 807–813
Wang Q, Liu F, Zhang DC (2020) Pelagihabitans pacificus gen. nov., sp. nov., a member of the family Flavobacteriaceae isolated from a deep-sea seamount. Int J Syst Evol Microbiol 70:4569–4575
Baik KS, Park SC, Kim HJ, Lee KH, Seong CN (2010) Chryseoglobus frigidaquae gen. nov., sp. nov., a novel member of the family Microbacteriaceae. Int J Syst Evol Microbiol 60:1311–1316
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Fitch WM (1971) Toward defining the course of evolution: minimum changes for a specific tree topology. Syst Zool 20:406–416
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Li R, Li Y, Kristiansen K, Wang J (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24:713–714
Li R, Zhu H, Ruan J, Qian W, Fang X et al (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272
Margesin R, Gander S, Zacke G, Gounot AM, Schinner F (2003) Hydrocarbon degradation and enzyme activities of cold-adapted bacteria and yeasts. Extremophiles 7:451–458
Dong XZ, Cai MY (eds) (2001) Determinative manual for routine bacteriology. Scientific Press, Beijing
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. Technical Note 101. Newark DE MIDI
Altenburger P, Kämpfer P, Makristathis A, Lubitz W, Busse H-J (1996) Classification of bacteria isolated from a medieval wall painting. J Biotechnol 47:39–52
Stolz A, Busse H-J, Kämpfer P (2007) Pseudomonas knackmussii sp. nov. Int J Syst Evol Microbiol 57:572–576
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917
Tindall BJ (1990) Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol Lett 66:199–202
Tindall BJ (1990) A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst Appl Microbiol 13:128–130
Schumann P (2011) Peptidoglycan structure. Method Microbiol 38:101–129
Schleifer KH (1985) Analysis of the chemical composition and primary structure of murein. Method Microbiol 18:123–156
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M et al (2012) Introducing EzTaxon: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR et al (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF et al (2011) Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinform 12:124
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282
Yoon SH, Ha SM, Lim J, Kwon S, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Yoon JH, Kang SJ, Schumann P, Oh TK (2006) Yonghaparkia alkaliphila gen. nov., sp. nov., a novel member of the family Microbacteriaceae isolated from an alkaline soil. Int J Syst Evol Microbiol 56:2415–2420
Tiago I, Morais PV, da Costa MS, Verissimo A (2006) Microcella alkaliphila sp. nov., a novel member of the family Microbacteriaceae isolated from a non-saline alkaline groundwater, and emended description of the genus Microcella. Int J Syst Evol Microbiol 56:2313–2316
Tiago I, Pires C, Mendes V, Morais PV, da Costa M, Verissimo A (2005) Microcella putealis gen. nov., sp. nov., a gram-positive alkaliphilic bacterium isolated from a nonsaline alkaline groundwater. Syst Appl Microbiol 28:479–487
Behrendt U, Schumann P, Hamada M, Suzuki K, Sproer C, Ulrich A (2011) Reclassification of Leifsonia ginseng (Qiu et al. 2007) as Herbiconiux ginsengi gen. nov., comb. nov. and description of Herbiconiux solani sp. nov., an actinobacterium associated with the phyllosphere of Solanum tuberosum L. Int J Syst Evol Microbiol 61:1039–1047
Kim BC, Park DS, Kim H, Oh HW, Lee KH et al (2012) Herbiconiux moechotypicola sp. nov., a xylanolytic bacterium isolated from the gut of hairy long-horned toad beetles, Moechotypa diphysis (Pascoe). Int J Syst Evol Microbiol 62:90–95
Hamada M, Komukai C, Tamura T, Evtushenko LI, Vinokurova NG et al (2012) Description of Herbiconiux flava sp. nov. and emended description of the genus Herbiconiux. Int J Syst Evol Microbiol 62:795–799
Acknowledgements
We are grateful for Center for Ocean Mega-Science (Chinese Academy of Sciences). We thank Dr Peter Schumann (DSMZ) for the analysis of the peptidoglycan structure.
Funding
This study was supported by the National Natural Science Foundation of China (31670002) and the Science & Technology Basic Resources Investigation Program of China (2017FY100804) and the Senior User Project of RV KEXUE (KEXUE2019G09).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. ZDC designed this research, QF and HWX isolated the strain and performed the initial cultivation and strain deposition, QF performed strain characterization, the electron microscopic and chemotaxonomic analysis, HWX performed the genomic and phylogenetic analysis, and QF and HWX drafted the manuscript. ZDC supervised the study and contributed to text preparation and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no competing interests.
Ethical Approval
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Qin, F., He, WX. & Zhang, DC. Marinisubtilis pacificus gen. nov., sp. nov., a Member of the Family Microbacteriaceae Isolated From a Deep-Sea Seamount. Curr Microbiol 78, 2136–2142 (2021). https://doi.org/10.1007/s00284-021-02468-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02468-y