Abstract
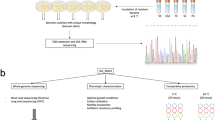
Psychrophiles, host of cold environments, have been successfully undergoing the process of evolution by which they have acquired innate adaptations to withstand the unfavorable effects of low temperature. Psychrophiles renders immense opportunity to explore the underlying mechanisms of cold adaptation. The present study focused to explore the cold adaptive mechanisms of Pseudomonas psychrophila MTCC12324, a facultative psychrophilic bacterium isolated from the Ny-Alesund, an island in the Svalbard Archipelago (79°55′ N, 11°56′ E) in the Arctic. Whole genome sequencing of P. psychrophila MTCC12324 and its analysis revealed the redundant nature of genome and identified several cold acclimation genes including cold shock proteins, and chaperones involved in the adaptive mechanism to thrive in the cold environment. Comparative proteome analysis of P. psychrophila MTCC12324 at 4 °C and 25 °C has thrown lights on the metabolic pathways and cellular processes adopted to withstand the cold environment. Basic survival pathways and factors involved in energy metabolism were found to be unaltered whereas stress response factors, enzymes involved in fatty acid elongation and cold-adapted chaperones were found to be enhanced towards cold stress. The present study facilitates recognition of crucial factors including polyunsaturated fatty acid biosynthesis, mRNA chaperones, and other cold-inducible proteins which favors the bacteria in conferring cold adaptation.



Similar content being viewed by others
Data Availability
The whole-genome shotgun sequence has been deposited in GenBank under the accession no. LBHT00000000. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD012212.
References
Russell NJ (2003) Psychrophily and resistance to low temperature. In: Gerday C, Glansdorff N (eds) Extremophiles, vol II. EOLSS, Paris
Gooch JW (2011) Facultative psychrophile. In: Gooch JW (ed) Encyclopedic dictionary of polymers. Springer, New York
Gounot AM, Russel NJ (1999) Physiology of cold-adapted microorganisms. In: Margesin R, Schinner F (eds) Cold-adapted organisms: ecology, physiology, enzymology and molecular biology. Springer, Berlin, pp 33–55
Price PB, Sowers T (2004) Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proc Natl Acad Sci USA 101:4631–4636. https://doi.org/10.1073/pnas.0400522101
Santelli CM, Orcutt BN, Banning E, Bach W, Moyer CL, Sogin ML et al (2008) Abundance and diversity of microbial life in ocean crust. Nature 453:653. https://doi.org/10.1038/nature06899
Methé BA, Nelson KE, Deming JW, Momen B, Melamud E, Zhang X et al (2005) The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc Natl Acad Sci USA 102:10913–10918. https://doi.org/10.1073/pnas.0504766102
Goodchild A, Saunders NF, Ertan H, Raftery M, Guilhaus M, Curmi PM, Cavicchioli R (2004) A proteomic determination of cold adaptation in the Antarctic archaeon, Methanococcoides burtonii. Mol microbiol 53:309–321. https://doi.org/10.1111/j.1365-2958.2004.04130.x
Allen MA, Lauro FM, Williams TJ, Burg D, Siddiqui KS, De Francisci D et al (2009) The genome sequence of the psychrophilic archaeon, Methanococcoides burtonii: the role of genome evolution in cold adaptation. ISME J 3:1012. https://doi.org/10.1038/ismej.2009.45
Nunn BL, Slattery KV, Cameron KA, Timmins-Schiffman E, Junge K (2015) Proteomics of Colwellia psychrerythraea at subzero temperatures: a life with limited movement, flexible membranes and vital DNA repair. Environ Microbiol 17:2319–2335. https://doi.org/10.1111/1462-2920.12691
Ma W, Jia J, Huang X, Xie W, Zhang X, Tang J, Lin C et al (2018) Stable isotope labelling by amino acids in cell culture (SILAC) applied to quantitative proteomics of Edwardsiella tarda ATCC 15947 under prolonged cold stress. Microb Pathog 125:12–19. https://doi.org/10.1016/j.micpath.2018.09.006
Radjasa OK, Nasima D, Sabdono A, Kita-Tsukamoto K, Ohwada K (2007) Characterization of psychrotrophic bacteria from sea waters of Makasar strait, Indonesia. J Biol Sci 7:658–662. https://doi.org/10.3923/jbs.2007.658.662
Abraham WP, Thomas S (2015) Draft genome sequence of Pseudomonas psychrophila MTCC 12324, isolated from the Arctic at 79 N. Genome Announc 3:e00578–e1515. https://doi.org/10.1128/genomeA.00578-15
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinform 10:421. https://doi.org/10.1186/1471-2105-10-421
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L et al (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. https://doi.org/10.1101/gr.089532.108
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genom 9:75. https://doi.org/10.1186/1471-2164-9-75
Yumoto I, Kusano T, Shingyo T, Nodasaka Y, Matsuyama H, Okuyama H (2001) Assignment of Pseudomonas sp. strain E-3 to Pseudomonas psychrophila sp. nov., a new facultatively psychrophilic bacterium. Extremophiles 5:343–349. https://doi.org/10.1007/s007920100199
Jiang B, Cui D, Li A, Gai Z, Ma F, Yang J, Ren N (2012) Genome sequence of a cold-adaptable sulfamethoxazole-degrading bacterium, Pseudomonas psychrophila HA-4. J Bacteriol 194(20):5721. https://doi.org/10.1128/JB.01377-12
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30(14):2068–2069. https://doi.org/10.1093/bioinformatics/btu153
Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J (2015) Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31(22):3691–3693. https://doi.org/10.1093/bioinformatics/btv421
Gopinath V, Raghunandanan S, Gomez RL, Jose L, Surendran A, Ramachandran R et al (2015) Profiling the proteome of Mycobacterium tuberculosis during dormancy and reactivation. Mol Cell Proteom 14:2160–2176. https://doi.org/10.1074/mcp.M115.051151
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Ayala-del-Río HL, Chain PS, Grzymski JJ, Ponder MA, Ivanova N, Bergholz PW et al (2010) The genome sequence of Psychrobacter arcticus 273–4, a psychroactive Siberian permafrost bacterium, reveals mechanisms for adaptation to low-temperature growth. Appl Environ Microbiol 76:2304–2312. https://doi.org/10.1128/AEM.02101-09
Bakermans C, Sloup RE, Zarka DG, Tiedje JM, Thomashow MF (2009) Development and use of genetic system to identify genes required for efficient low-temperature growth of Psychrobacter arcticus 273–4. Extremophiles 13:21–30. https://doi.org/10.1007/s00792-008-0193-3
Yamanaka K, Fang L, Inouye M (1998) The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol microbiol 27:247–255. https://doi.org/10.1046/j.1365-2958.1998.00683.x
Smith AL, Skerlos SJ, Raskin L (2013) Psychrophilic anaerobic membrane bioreactor treatment of domestic wastewater. Water Res 47:1655–1665. https://doi.org/10.1016/j.watres.2012.12.028
Zheng S, Ponder MA, Shih JYJ, Tiedje JM, Thomashow MF, Lubman DM (2007) A proteomic analysis of Psychrobacter articus 273-4 adaptation to low temperature and salinity using a 2-D liquid mapping approach. Electrophoresis 28:467–488. https://doi.org/10.1002/elps.200600173
Suryaletha K, Narendrakumar L, John J, Radhakrishnan MP, George S, Thomas S (2019) Decoding the proteomic changes involved in the biofilm formation of Enterococcus faecalis SK460 to elucidate potential biofilm determinants. BMC Microbiol 19:146. https://doi.org/10.1186/s12866-019-1527-2
Jones PG, Inouye M (1994) The cold-shock response: a hot topic. Mol Microbiol 11:811–818. https://doi.org/10.1111/j.1365-2958.1994.tb00359.x
Feller G, Gerday C (2003) Psychrophilic enzymes: hot topics in cold adaptation. Nat Rev Microbiol 1:200. https://doi.org/10.1038/nrmicro773
Ting L, Williams TJ, Cowley MJ, Lauro FM, Guilhaus M, Raftery MJ, Cavicchioli R (2010) Cold adaptation in the marine bacterium, Sphingopyxis alaskensis, assessed using quantitative proteomics. Environ Microbiol 12:2658–2676. https://doi.org/10.1111/j.1462-2920.2010.02235.x
D'Amico S, Collins T, Marx JC, Feller G, Gerday C (2006) Psychrophilic microorganisms: challenges for life. EMBO Rep 7:385–389. https://doi.org/10.1038/sj.embor.7400662
Saunders NF, Thomas T, Curmi PM, Mattick JS, Kuczek E, Slade R et al (2003) Mechanisms of thermal adaptation revealed from the genomes of the Antarctic Archaea Methanogenium frigidum and Methanococcoides burtonii. Genome Res 13:1580–1588. https://doi.org/10.1101/gr.1180903
Metpally RPR, Reddy BVB (2009) Comparative proteome analysis of psychrophilic versus mesophilic bacterial species: insights into the molecular basis of cold adaptation of proteins. BMC Genom 10:11. https://doi.org/10.1186/1471-2164-10-11
Russell NJ (1997) Psychrophilic bacteria-molecular adaptations of membrane lipids. Comp Biochem Physiol A 118:489–493. https://doi.org/10.1016/S0300-9629(97)87354-9
García-Arribas O, Mateo R, Tomczak MM, Davies PL, Mateu MG (2007) Thermodynamic stability of a cold-adapted protein, type III antifreeze protein, and energetic contribution of salt bridges. Protein Sci 16:227–238. https://doi.org/10.1110/ps.062448907
Thieringer HA, Jones PG, Inouye M (1998) Cold shock and adaptation. BioEssays 20:49–57. https://doi.org/10.1002/(SICI)1521-1878(199801)20:1%3c49:AID-BIES8%3e3.0.CO;2-N
Broeze RJ, Solomon CJ, Pope DH (1978) Effects of low temperature on in vivo and in vitro protein synthesis in Escherichia coli and Pseudomonas fluorescens. J Bacteriol 134:861–874
Fang L, Jiang W, Bae W, Inouye M (1997) Promoter-independent cold-shock induction of cspA and its derepression at 37° C by mRNA stabilization. Mol Microbiol 23:355–364. https://doi.org/10.1046/j.1365-2958.1997.2351592.x
Jones PG, Inouye M (1996) RbfA, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol Microbiol 21:1207–1218. https://doi.org/10.1111/j.1365-2958.1996.tb02582.x
Suzuki I, Kanesaki Y, Mikami K, Kanehisa M, Murata N (2001) Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol Microbiol 40:235–244. https://doi.org/10.1046/j.1365-2958.2001.02379.x
Acknowledgements
ST acknowledges Ministry of Earth Sciences, Govt. of India for all the financial and logistic support during Indian Arctic Expedition 2009, Dr. Rasik Ravindra, former Director of The National Centre for Polar and Ocean Research (NCPOR), Goa and Dr. K.P. Krishnan, other scientists and staff of NCPOR for the necessary help during Indian Arctic Expedition. We also acknowledge Dr. Abdul Jaleel K.A, Aneesh Kumar A and Arun Surendran at the Proteomics Core Facility for LC–MS/MS analysis. The authors thank Sivakumar K.C for the timely help in pan-genome analysis. The authors would like to thank Prof. M. Radhakrishna Pillai, Director, Rajiv Gandhi Centre for Biotechnology for providing the necessary facilities.
Author information
Authors and Affiliations
Contributions
Conceptualization-WPA and ST; Methodology- WPA, SR, VG, and ST; Data analysis-WPA, SR, and ST; Manuscript drafting and editing-WPA, SR, KS and ST.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
284_2020_2006_MOESM1_ESM.xls
Table S1. List of genes common among the test strain (P. psychrophila MTCC 12,324) and either of the reference strains (P. psychrophila HA-4/ P. psychrophila JSM 10,889). Supplementary file1 (XLS 239 kb)
284_2020_2006_MOESM2_ESM.doc
Table S2: Genome to Genome comparison between Pseudomonas psychrophila MTCC12324 & Psychrobacter arcticus 273-4. Supplementary file2 (DOC 157 kb)
284_2020_2006_MOESM3_ESM.docx
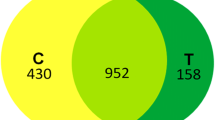
Table S3: List of total proteins of P. psychrophila MTCC12324 and their expression at 4 °C which fall in biological processes such as (A) DNA replication, repair, recombination & cell division (B) Transcription and RNA processing (C) Translation (D) Protein folding and processing (E) Carbohydrate and energy metabolism (F) Amino acid metabolism (G) Nucleotide metabolism (H) Fatty acid metabolism (I) Transport. A fold change higher than 70% (ratio of either < 0.70 for downregulation or > 1.3 for upregulation) was considered to be indicative of significantly altered levels of expression.4 °C & 25 °C indicates the unique representation of corresponding proteins in their respective conditions. Supplementary file3 (DOCX 123 kb)
Rights and permissions
About this article
Cite this article
Abraham, W.P., Raghunandanan, S., Gopinath, V. et al. Deciphering the Cold Adaptive Mechanisms in Pseudomonas psychrophila MTCC12324 Isolated from the Arctic at 79° N. Curr Microbiol 77, 2345–2355 (2020). https://doi.org/10.1007/s00284-020-02006-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02006-2